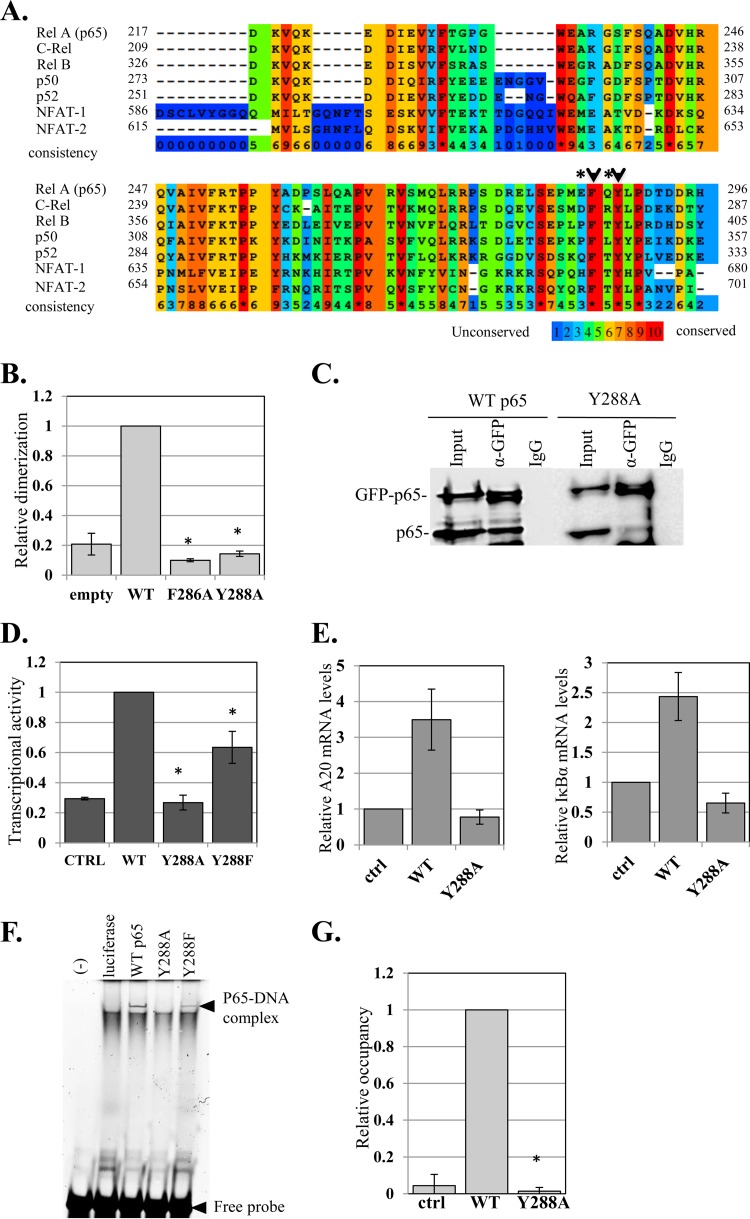
FIG 5.
The effects of the nonconserved E285 and Q287 on dimerization are linked to the highly conserved F286 and Y288. (A) Amino acid sequence alignment of five NF-κB and two NFAT proteins. Surface residues E285 and Q287 are marked by asterisks and adjacent HCD residues with arrowheads. (B) F286 and Y288, which are adjacent to E285 and Q287, are essential for dimerization. Shown is a split-RL dimerization assay using WT p65 and F286A and Y288A mutants. (C) p65-Y288A mutation impairs dimerization. Cells were cotransfected with p65 and GFP-p65 (both WT and the Y288A mutant) and harvested 48 h later. The lysates were immunoprecipitated either with anti-GFP antibody or with a control antibody. The input (10%) and the immune complexes were analyzed by Western blotting using anti-p65 antibody. The lanes of the Y288A immunoprecipitates (IP) were spliced to bring them close to the WT lanes. The original gel is shown in Fig. S4 in the supplemental material. (D) Activation of the A20 promoter-luciferase by WT, Y288A, and Y288F p65 variants. The bars represent the means and SE of the results of 3 independent experiments. (E) HEK293T cells were transfected with p65 WT and Y288A, and 24 h later, total RNA was extracted. The levels of A20, IκBα, and GAPDH mRNAs were determined by RT-qPCR. The bars represent the means ± SD of A20 and IκBα levels normalized to GAPDH of 2 independent experiments. (F) Y288 mutations impair DNA binding. The WT and Y288A and Y288F p65 variants were synthesized in vitro in rabbit reticulocyte lysate and then subjected to EMSA using a fluorescently labeled DNA probe. The positions of the p65-DNA complex and the free DNA are indicated. (G) HEK293T cells were transfected with p65 WT and Y288A and 24 h later subjected to ChIP using anti-p65 or control (for background levels) antibodies. Analysis was performed by qPCR. The graph shows occupancy levels normalized to input levels. The uninduced sample was set to 1. The results represent the averages ± SEM of the results of at least 3 independent experiments. The asterisks denote statistically significance differences (P < 0.05).