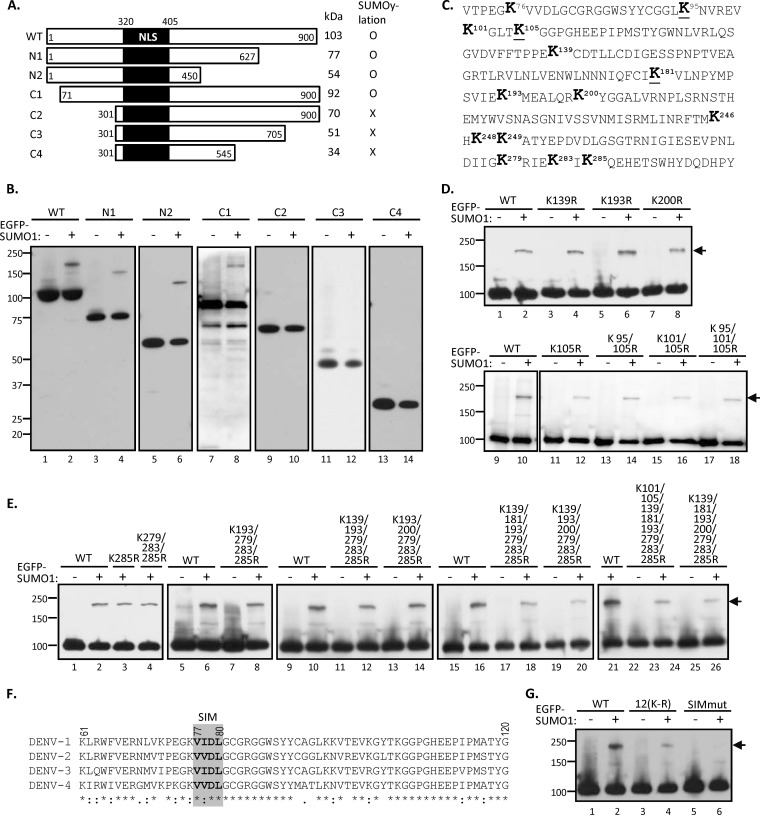
FIG 3.
Putative SUMO-interacting motif (SIM) located in the N-terminal domain of NS5 is crucial for NS5 SUMOylation. (A) Schematic diagram of NS5 and its truncation derivatives. The predicted sizes and the ability of NS5 derivatives to undergo SUMOylation are summarized on the right. NLS, nuclear localization signal. (B) HEK293T cells were cotransfected with Ubc9 and each NS5 construct with (+) or without (−) EGFP-tagged SUMO1 for 48 h. Transfectants were harvested and analyzed by IP-WB using anti-HA antibody. (C) Amino acid sequence of the N-terminal 71 to 300 residues in DENV2 NS5. The lysine (K) residues are shown in boldface, whereas the conserved K residues among four serotypes of DENV are underlined. (D) In HEK293T cells, plasmid expressing NS5-WT or the indicated mutants containing single, double, or triple K-R substitution(s) were cotransfected with or without EGFP-SUMO1 and Ubc9. The whole-cell extracts then were collected and subjected to IP-WB analysis by anti-HA antibody. (E) HEK293T cells were cotransfected with Ubc9, EGFP-SUMO1, and each NS5 construct containing multiple K-R substitutions as indicated for 48 h. Transfectants were harvested and analyzed by IP-WB using anti-HA antibody. (F) Protein alignment of the N-terminal sequences from four different serotypes of DENV NS5. The putative SIM of DENV NS5 is shown in the gray square. (G) HEK293T cells were cotransfected with Ubc9 and/or EGFP-SUMO1 plus NS5-WT or its mutant containing 12 K-R substitutions [termed 12(K-R)] or VVDL-to-AAAA substitution at the putative SIM motif (SIMmut). Cells were harvested at 48 h posttransfection for IP-WB analysis using anti-HA antibody. Arrow, SUMOylated NS5 proteins.