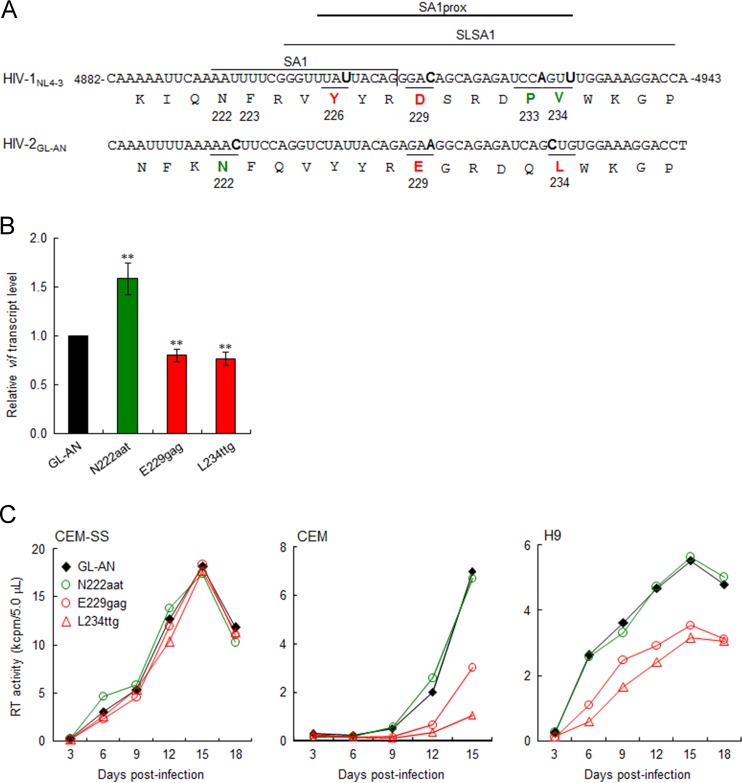
FIG 3.
Effects of natural single-nucleotide variations in HIV-2 SA1prox on vif transcript levels and viral replication ability. Red and green represent low and high expressers of the vif transcript, respectively. (A) Sequences of HIV-1NL4-3 (nucleotides 4882 to 4943 and Pol-IN amino acids 219 to 238) and of the corresponding region of HIV-2GL-AN. SA1 (7–10), SLSA1 (54), and SA1prox (6) in the NL4-3 sequences are shown. Letters in boldface type in the sequences represent the sites at which natural variations that alter virus replication ability (NL4-3 [6] and GL-AN [this study]) are located. (B) vif transcript levels for GL-AN and its natural variants. The proviral clones indicated were transfected into HEK 293T cells, and at 20 h posttransfection, total RNAs were prepared for qRT-PCR analysis using a primer pair specific for the GL-AN vif transcript. vif transcript levels relative to those of GL-AN are presented. Mean values ± standard deviations from three independent experiments are shown. Significance relative to GL-AN was determined by the Student t test (**, P < 0.05). (C) Viral growth kinetics of GL-AN and its natural variants in CEM-SS, CEM, and H9 cells. Viruses were prepared from transfected HEK 293T cells, and 105 cells were infected with viruses (2 × 105 RT units for CEM-SS and CEM cells and 5 × 104 RT units for H9 cells). Virus replication was monitored by the activity of RT released into the culture supernatants. Representative data from at least two independent experiments are shown.