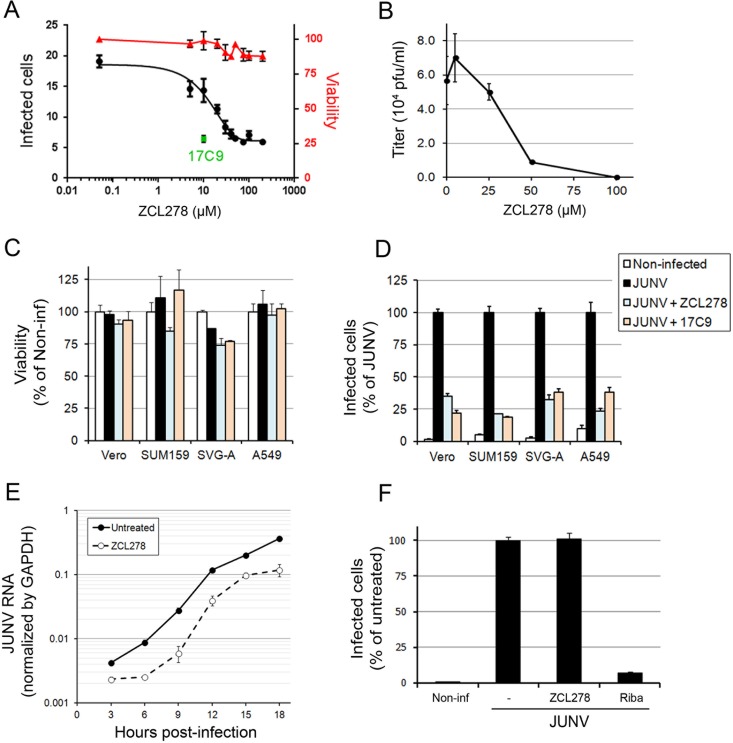
FIG 3.
Identification of ZCL278 as a potent JUNV entry inhibitor. (A) Cells were untreated or pretreated for 15 min with ZCL278 and subsequently infected with JUNV (MOI, 0.3) in the presence or absence of the compound for an additional hour. Then, the cells were washed and incubated for 16 h in virus medium containing an anti-GP neutralizing antibody (GD01). Cells were fixed and stained with an anti-NP antibody coupled to A647, and the percentage of infected cells was measured by flow cytometry. The percentage of infected Vero cells (left axis) was plotted against the ZCL278 concentration (black line). Samples treated in parallel were used to measure cell viability using the CellTiter-Glo assay. Viability (right axis), which was normalized to that of the untreated sample, is shown by the red line. 17C9 at a single concentration of 10 μM was used as a positive control, and the percentage of infected cells is shown as a green dot. Error bars are the means ± SDs from duplicate experiments. (B) Vero cells were untreated or pretreated with increasing amounts of ZCL278 for 15 min and subsequently infected with JUNV for 1 h in the presence or absence of ZCL278. After 24 h, the supernatant was harvested and the number of PFU of virus per milliliter was measured by plaque assay. Error bars are the means ± SDs from duplicate experiments. (C, D) Vero, SUM159, SVG-A, or A549 cells were treated as described in the legend to panel A. (C) Viability was determined by measuring the luminescence using the CellTiter-Glo assay. Error bars are the means ± SDs from triplicate experiments. (D) The normalized percentage of infected cells of each cell type and condition. Error bars are the means ± SDs from duplicate experiments with at least 10,000 cells per condition. (E) One-step growth curve of Vero cells that were infected with JUNV at an MOI of 3 and that were pre- and cotreated with 50 μM ZCL278 as described in the legend to panel A. RNAs were extracted at the indicated times postinfection. An RT-qPCR specific for JUNV RNA was performed, and values were normalized by GAPDH values. Error bars are the means ± SDs from duplicate experiments. The differences between untreated and ZCL278-treated cells were highly significant at every time point. (F) Vero cells were first infected with JUNV for 1 h and subsequently incubated with 50 μM ZCL278 or 400 μM ribavirin (Riba) in virus medium containing an anti-GP neutralizing antibody for 15 h. Cells were fixed, stained, and analyzed by flow cytometry as described in the legend to panel A. The data were normalized to the percentage of untreated infected cells. Error bars are the means ± SDs from duplicate experiments with at least 10,000 cells per condition.