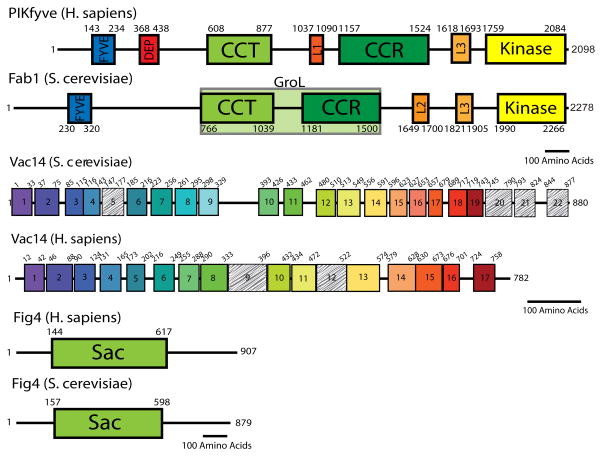
Figure 2. Domain architecture of Fab1/PIKfyve, Vac14, and Fig4.
Boundaries of each domain were determined using a combination of Jpred4 secondary structure prediction and ClustalW multiple sequence alignment [72, 73]. For Vac14, the above techniques were used in addition to tailored HHpred alignments of select predicted HEAT repeats [74]. Fab1/PIKfyve contains previously described domains (FYVE, DEP, CCT, CCR, and Kinase); we identify three additional areas of predicted secondary structure which have structural and sequence conservation in all species (L3), in all fungi (L2), or in metazoans (L1). Vac14 is composed of tandem HEAT repeats. Colored boxes indicate homology of HEAT repeats between yeast and human Vac14. Hashed boxes indicate degenerate sequences which may be bona fide HEAT repeats. Fig4 contains a single Sac domain, which is conserved in some lipid phosphatases. Black lines represent 100 amino acids.