Abstract
Removal of the 5′-end 7-methylguanosine cap structure is a critical step in the highly regulated process of mRNA decay. The Nudix hydrolase, Dcp2, was identified as a first decapping enzyme and subsequently shown to preferentially modulate stability of only a subset of mRNAs. This observation led to the hypothesis that mammalian cells possess multiple decapping enzymes that may function in distinct pathways. Here we report Nudt3 is a Nudix protein that possesses mRNA decapping activity in cells and is a modulator of MCF-7 breast cancer cell migration. Reduction of Nudt3 protein levels in MCF-7 cells promotes increased cell migration and corresponding enhanced filopodia extensions. Importantly, this phenotype was reversed by complementation with wild type, but not catalytically inactive Nudt3 protein indicating Nudt3 decapping activity normally functions to control cell migration. Genome-wide analysis of Nudt3 compromised cells identified elevated levels of transcripts involved in cell motility including integrin β6, lipocalin-2, and fibronectin. The observed increase in mRNA abundance was dependent on Nudt3 decapping activity where integrin β6 and lipocalin-2 were modulated directly through mRNA stability, while fibronectin was indirectly controlled. Moreover, increased cell migration observed in Nudt3 knockdown cells was mediated through the extracellular integrin β6 and fibronectin protein nexus. We conclude that Nudt3 is an mRNA decapping enzyme that orchestrates expression of a subset of mRNAs to modulate cell migration and further substantiates the existence of multiple decapping enzymes functioning in distinct cellular pathways in mammals.
Keywords: Nudt3, mRNA decapping, mRNA stability, cell motility, integrin β6, fibronectin
INTRODUCTION
All eukaryotic cellular mRNAs possess distinctive features at their 5′- and 3′-ends. The presence of 5′ 7-methyloguanosine cap structure and the 3′-terminal poly(A) tract fulfill a variety of functions in synthesis, translation, and degradation of mRNA. These two elements act cooperatively and in close proximity to protect mRNA against exonucleolytic degradation (Furuichi et al. 1977) and synergistically enhance mRNA translation (Gallie 1991; Preiss and Hentze 1998). The decay of mRNA is highly regulated, and in mammalian cells it starts with an initial deadenylation step to remove the poly(A) tail (Chen and Shyu 2011). The remaining mRNA is either degraded by 5′–3′ or 3′–5′ pathway or undergoes endonucleolytic or enzymatic decay by specialized routes for aberrant mRNA degradation (Coller and Parker 2004). In the 5′–3′ decay pathway, removal of the cap structure occurs rapidly after poly(A) tract shortening (Muhlrad et al. 1995). This process is carried out by the decapping enzymes, which in turn exposes the transcripts to digestion by a highly processive 5′–3′-exonuclease, Xrn1 (Hsu and Stevens 1993).
Dcp2, a member of the Nudix family of pyrophosphatases, was identified as the first mammalian decapping enzyme (Lykke-Andersen 2002; van Dijk et al. 2002; Wang et al. 2002). Dcp2 was subsequently shown to be developmentally regulated, not expressed ubiquitously in all tissues and primarily functions on a subset of mRNAs (Li et al. 2008, 2011, 2012; Song et al. 2010). The mammalian Nudt16 was the second mammalian decapping enzyme identified and despite its ubiquitous expression, also primarily targets a subset of mRNAs (Song et al. 2010) suggesting additional uncharacterized decapping enzymes are functional in mammalian cells.
Dcp2 (Nudt20) and Nud16 are both members of the Nudix hydrolase superfamily of proteins that are known to catalyze hydrolysis of a wide range of small nucleotide substrates composed of a nucleoside diphosphate linked to another moiety X (Nudix) (Bessman et al. 1996). They contain a conserved Nudix motif with the consensus sequence GX5EX7REUXEEXGU (where U represents a hydrophobic residue, and X represents any amino acid), which forms part of the versatile catalytic site for diphosphate hydrolysis (Bessman et al. 1996). The Nudix family of proteins are evolutionarily conserved with 22 Nudix proteins identified in the human genome. Although Dcp2 and Nudt16 are the only known mammalian Nudix proteins that have been reported to decap RNA in cells, six additional mammalian Nudix proteins possess decapping activity at least in vitro (Song et al. 2013). Nudt2, Nudt3, Nudt12, and Nudt15 hydrolyze methylated capped RNA with the activity comparable to Dcp2 or Nudt16, while Nudt17 and Nudt19 possess modest decapping activity in vitro (Song et al. 2013). Six Nudix proteins are present in yeast Saccharomyces cerevisiae. However, other than Dcp2, only one of the five Nudix proteins, Ddp1p, demonstrated robust decapping activity in vitro (Song et al. 2013). Interestingly, Ddp1p appears to be the homolog of mammalian Nudt3 suggesting that Nudt3 and Ddp1p may be evolutionarily conserved decapping enzymes.
Nudt3 is a cytoplasmic protein that, along with Nudt4, 10, and 11 belong to the diphosphoinositol polyphosphate phosphohydrolases (DIPP) family of proteins and shown to hydrolyze diphosphoinositol polyphosphates (DIPs) and ApnA dinucleotide (McLennan 2006). Nudt3 is ubiquitously expressed in mouse tissue (Williams et al. 2015). Polymorphism of NUDT3, among more than 50 genetic variants, was implicated by genome-wide association to increase BMI and obesity (Hofker and Wijmenga 2009) possibly through its phosphatase activity against polyphosphate substrates (Shears 2009). In this report, we provide several lines of evidence that Nudt3 is a decapping enzyme in cells and demonstrate that Nudt3 regulates MCF-7 cell motility by directly affecting stability of mRNAs involved in cellular migration.
RESULTS
Steady-state levels of mRNAs implicated in cellular movement are increased in Nudt3 knockdown cells
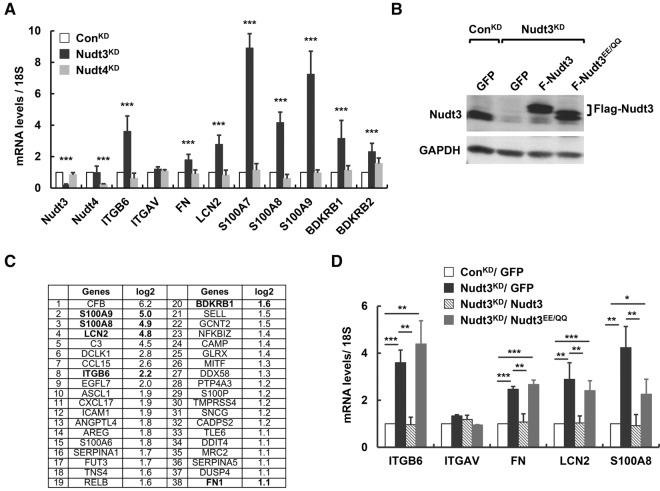
Our recent demonstration that both mammalian Nudt3 and its yeast homolog, Ddp1p, possess in vitro decapping activity prompted us to test whether Nudt3 is a decapping enzyme in cells. MCF-7 cells constitutively expressing either a short hairpin RNA (shRNA) directed against Nudt3 (Nudt3KD) or a nontargeted control shRNA (ConKD) were used. Nudt3 mRNA was reduced by 80% with a corresponding significant reduction of the protein levels in Nudt3KD cells (Fig. 1A,B). A genome-wide RNA-seq analysis was carried out to comprehensively identify transcripts whose steady-state levels were selectively altered by Nudt3. RNA from Nudt3KD and ConKD cells (n = 3 samples per group) were subjected to high-throughput sequencing using Illumina sequencing technology. Sequencing results were aligned to the human genome and compared using standard bioinformatics methods (Trapnell et al. 2013). Comparison of RNA profiles from ConKD and Nudt3KD cells revealed that steady-state levels of 161 RNA were altered more than twofold between the two cell lines with false discovery rates (FDR) <0.05. In this group, we found 144 and 17 RNAs to be up-, or down-regulated, respectively. Interestingly, subjecting the increased RNAs to gene ontology analyses algorithms within Ingenuity Pathway Analysis (IPA) revealed a group of 38 transcripts potentially involved in cellular movement (Fig. 1C).
FIGURE 1.
Nudt3 down-regulation increases expression of mRNAs implicated in cell motility. (A) RNA levels of nine mRNAs implicated in cell migration tested in MCF-7 cells expressing nontargeting shRNA (ConKD, open bar), shRNA against Nudt3 (Nudt3KD, filled bar), or shRNA against Nudt4 (Nudt4KD, gray bar) are shown. (B) Western blot analysis of Nudt3 protein levels in ConKD or Nudt3KD MCF-7 cells ectopically expressing GFP, Flag-tagged wild-type Nudt3 (Nudt3), or mutant Nudt3 (Nudt3EE/QQ). The band for Flag-Nudt3EE/QQ runs slightly faster than that for Flag-Nudt3 likely due to differences in protein charge by the introduction of glutamic acid residues into the mutant protein. (C) A subset of the mRNAs implicated in cellular movement derived from the 144 mRNAs detected to increase greater than or equal to twofold by RNA-seq analysis in Nudt3KD are shown. (D) Transcript levels of the indicated mRNAs were determined by qRT-PCR in ConKD or Nudt3KD cells complemented with the indicated proteins. RNA levels in A and D are derived from at least three independent experiments and normalized to 18S rRNA and presented relative to the value in control cells set to one. Error bars represent ±SD. P-values are denoted by asterisks. (*) P < 0.05, (**) P < 0.01, (***) P < 0.001 (Student's t-test).
A subset of nine RNAs implicated in cellular migration were chosen for further validation by direct quantitative reverse-transcription (qRT)-PCR. Steady-state levels of eight were found reproducibly and significantly elevated more than twofold in Nudt3KD compared with ConKD cells (Fig. 1A). The catalytic activity of Nudt3 has been shown to hydrolyze two primary substrates in vitro, dinucleoside polyphosphates (ApnA) (McLennan 2006) and capped mRNA (Song et al. 2013). To distinguish which of these activities contributed to Nudt3's role in up-regulating specific mRNA levels, we determined the mRNA expression in cells lacking Nudt4. Nudt3 and Nudt4 both hydrolyze dinucleoside polyphosphates as well as nonnucleotide substrates such as diphosphoinositol polyphosphates (DIPs) (McLennan 2006). However, only Nudt3 possesses RNA-decapping activity as demonstrated by in vitro decapping assays. MCF-7 cell lines constitutively expressing a Nudt4-directed shRNA, which reduced Nudt4 mRNA levels by 80% (Nudt4KD) were generated (Fig. 1A). The steady-state level of mRNAs whose expressions were responsive to Nudt3 deficiency remained unchanged in Nudt4KD cells (Fig. 1A), suggesting that the elevated mRNAs levels implicated in cellular movement were not mediated through common dinucleotide substrates and likely through the mRNA decapping activity of Nudt3.
To determine whether the catalytic activity of Nudt3 was necessary, Nudt3KD cells were complemented with Flag-tagged shRNA-resistant wild-type or a catalytically inactive mutant (Nudt3EE/QQ) harboring glutamine substitutions for two glutamic acid metal coordination residues that abolish Nudt3 decapping activity (Song et al. 2013). Exogenous proteins were expressed at levels comparable to endogenous Nudt3 (Fig. 1B). Elevated levels of integrin β6 (ITGB6), fibronectin (FN), lipocalin-2 (LCN2), and S100 calcium-binding protein A8 (S100A8) mRNAs in Nudt3-deficient cells were complemented by wild-type Nudt3, but not by the decapping-deficient Nudt3EE/QQ (Fig. 1D). These data demonstrated that the observed steady-state mRNA increases were a consequence of the Nudt3 catalytic activity.
Nudt3 is a decapping enzyme that modulates the stability of a subset of mRNAs implicated in cell motility
To directly test whether Nudt3 modulates the stability of mRNA and functions as a decapping enzyme, the stability of four select mRNAs elevated in the Nudt3KD cells and integral to cell motility were tested. RNA half-lives for ITGB6, LCN2, FN, and S100A8 were determined following actinomycin D-directed transcriptional silencing. Consistent with Nudt3 functioning as a decapping enzyme in cells, the stability of ITGB6 and LCN2 mRNAs were increased in Nudt3KD compared with ConKD cells (Fig. 2). Importantly, mRNA stabilization was exclusively reversed by complementation with wild-type Nudt3 but not Nudt3 harboring the catalytic site mutations, demonstrating that these two transcripts are direct targets of the Nudt3 decapping activity. Surprisingly, stability of FN and S100A8 mRNAs were unaltered irrespective of Nudt3 levels, indicating that changes in the steady-state levels of these mRNAs observed in Figure 1D were an indirect consequence of Nudt3 and not by direct decapping of either mRNA. Collectively, our data revealed that Nudt3 functions as a decapping enzyme in cells by modulating the stability of a subset of mRNAs.
FIGURE 2.
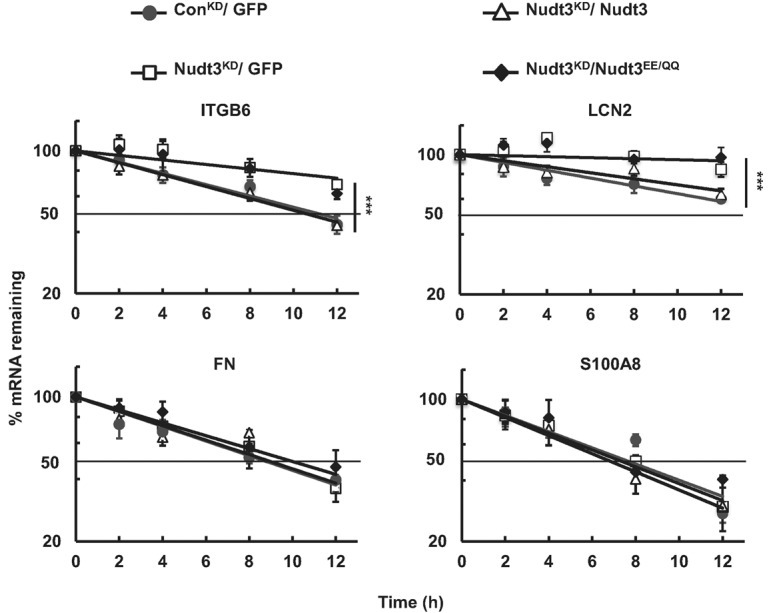
Nudt3 is a decapping enzyme that modulates stability of integrin β6 and lipocalin-2 mRNAs. Stability of the indicated mRNAs was determined following actinomycin D-mediated transcriptional silencing in ConKD and Nudt3KD cells complemented with the indicated proteins. Values are normalized to levels of 18S rRNA and presented relative to their respective levels at time zero. Data were derived from at least three independent experiments. The dotted line demarcates 50%. P-values from comparison of the decay rates are presented with asterisks. (***) P < 0.01 (two-tailed extra sum-of-squares F-test).
Nudt3 down-regulation alters cell morphology and increases motility of MCF-7 cells
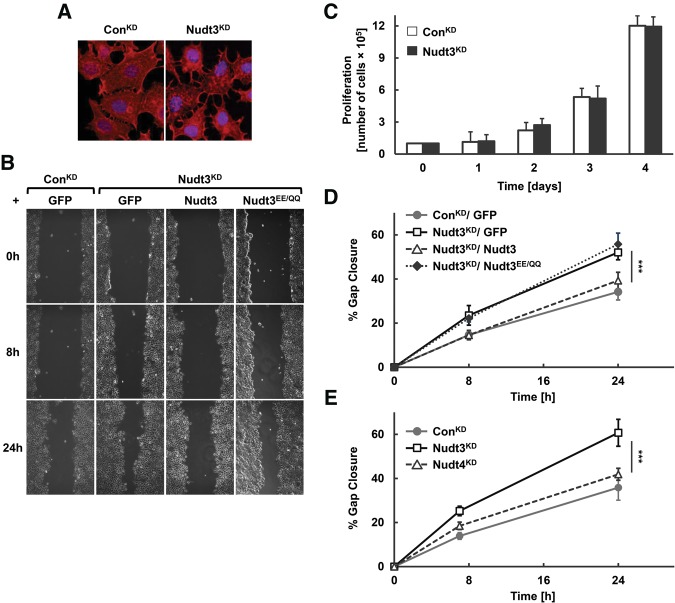
Examination of Nudt3KD cells revealed an interesting altered cell morphology consistent with a migratory phenotype variance. Fluorescence staining of actin protein showed enhanced lamellipodia/filopodia extensions in MCF-7 cells knocked down for Nudt3 (Nudt3KD) (Fig. 3A) compared with control cells, suggesting an increase in cell migration. We next examined whether a reduction of Nudt3 affects the motile properties of MCF-7 cells with an in vitro scratch-wound healing assay (Liang et al. 2007). In this assay, cells migrate to close an initial gap made after a “scratch” in a cell monolayer. MCF-7 cells stably expressing Nudt3 shRNA had significantly increased motility compared with control shRNA cells (Fig. 3B). The increased motility of Nudt3KD cells was not a function of an increased proliferation rate since the studies were carried out in the presence of mitomycin C and further confirmed by the lack of any significant difference between ConKD and Nudt3KD cell proliferation (Fig. 3C). To determine whether the migratory effects were directly related to the loss of Nudt3 decapping activity, Nudt3KD cells were complemented with shRNA-resistant Flag-tagged Nudt3 or the Nudt3EE/QQ mutant. Nudt3KD MCF-7 cells were markedly less motile upon expression of Nudt3 but not Nudt3EE/QQ (Fig. 3B,D), indicating that the increased cell motility in Nudt3KD cells was a consequence of reduced Nudt3 decapping activity. Moreover, Nudt4KD cells displayed migration undistinguishable from ConKD cells (Fig. 3E) further emphasizing Nudt3 decapping activity for the migratory phenotype. Collectively, the increased motility of Nudt3 knockdown cells was due to a lack of Nudt3 catalytic activity and likely a consequence of inhibiting its mRNA decapping activity.
FIGURE 3.
Nudt3 is involved in reducing MCF-7 cell migration. (A) Immunofluorescence of filamentous (F)-actin (red) and DNA (blue) in proliferating ConKD and Nudt3KD MCF-7 cells. (B) Motility of ConKD or Nudt3KD MCF-7 cells complemented with the indicated proteins were analyzed by an in vitro scratch-wound healing assay. The cells were cultured to confluent cell monolayers, treated with mitomycin C to inhibit cell division, scratched and photographed at 0, 8, and 24 h after wounding. (C) Proliferation of ConKD and Nudt3KD MCF-7 cells was quantified by counting the number of cells at day 1, 2, 3, and 4 after initial seeding of 1 × 105 cells per 35-mm dish. Data are representative of three independent experiments. (D,E) The percentage of gap closure for Nudt3KD and Nudt4KD cells were quantitated with ImageJ software. At least five different wounds were quantified for each time point. Data are representative of three independent experiments ±SD. P-values are denoted by asterisks. (***) P < 0.001 (Student's t-test).
Nudt3 influences cell migration through the integrin β6-fibronectin nexus
The molecular pathway by which Nudt3 influences cell migration was next addressed. We initially focused on ITGB6 and the stromal extracellular matrix (ECM) protein, FN, whose mRNAs were both significantly altered in a Nudt3-dependent manner and contribute to cell migration (Huang et al. 1998; Ridgway et al. 2005; Pereira et al. 2014). FN is an important ligand in promoting cell adhesion and migration (Weinacker et al. 1994) and interacts with multiple cell surface integrin receptors including the αvβ6 heterodimer, which enables cellular communication with the ECM (Humphries et al. 2006). αvβ6 consists of an αv and the β6 encoded by ITGB6. Although levels of the αv mRNA component of the heterodimer were similar between Nudt3KD and ConKD cell lines, the ITGB6 mRNA was elevated fourfold in Nudt3KD cells (Fig. 1B). αvβ6 expression is minimal to negligible in normal epithelium but highly up-regulated in neoplastic epithelial cells (Breuss et al. 1995; Bandyopadhyay and Raghavan 2009). Consistent with the mRNA data, ITGB6 protein was markedly increased in Nudt3KD cell extracts but, as expected, not detected in ConKD cells (Fig. 4A).
FIGURE 4.
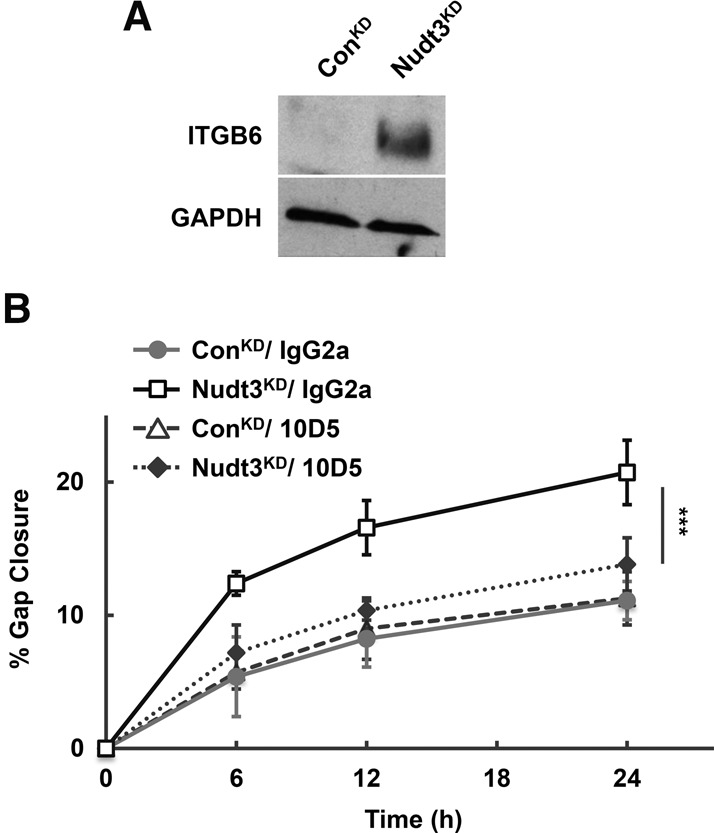
Elevated ITGB6 expression in Nudt3 knockdown cells is essential for increased MCF-7 cells motility. (A) A representative Western blot of integrin β6 (ITGB6) protein in cell extract from ConKD and Nudt3KD MCF-7 cells is shown. (B) Cell migration of ConKD and Nudt3KD MCF-7 cells in the presence of 10 μg/mL of either αvβ6-neutralizing antibody (10D5) or nonspecific antibody (IgG2a) in the medium lacking FBS was assessed by scratch-wound healing assays. At least five different wounds were quantified for each time point. Data are representative of three independent experiments ±SD. P-values are denoted by asterisks. (***) P < 0.001 (Student's t-test).
To determine whether integrin αvβ6 is involved in Nudt3-mediated cell migration, we utilized the αvβ6 neutralizing antibody 10D5 (Munger et al. 1999). As expected, Nudt3KD cells demonstrated approximately twofold increase in cell motility compared to ConKD in the presence of nonspecific IgG2a antibody (Fig. 4B). Interestingly, the motility of the Nudt3KD cells was decreased to the same level as ConKD cells only in the presence of αvβ6-specific, 10D5 antibody but not by the nonspecific IgG2a antibody (Fig. 4B). We observed similar cell motility for ConKD cells cultured either in the presence of IgG2a or 10D5 antibody which correlates with negligible αvβ6 expression in normal epithelium. These results indicate that integrin αvβ6 plays a crucial role in the Nudt3-mediated increase in cell motility likely through its modulation of ITGB6 mRNA.
The significance of Nudt3-mediated regulation of FN was next addressed. To determine whether elevation of FN mRNA could contribute to the enhanced motility phenotype observed in the Nudt3KD cells, levels of secreted FN protein were tested in Nudt3KD cultures. As show in Figure 5A,B, a dramatic increase of FN protein was detected in Nudt3KD cell culture media consistent with the increased FN mRNA in these cells (Fig. 1B). Moreover, the increase was reversed by complementation by exogenous wild-type Flag-Nudt3 (Fig. 5B, lane 2 versus 3), but not the catalytically inactive mutant Flag-Nudt3EE/QQ (lane 2 versus 4) establishing a role for Nudt3 decapping in the regulation of secreted FN protein levels. Importantly, MCF-7 cells compromised in both FN and Nudt3 expression (Fig. 5A, lanes 3 and 4) failed to exhibit the increased motility observed in cells depleted of Nudt3 alone and displayed a migratory phenotype comparable to ConKD cells (Fig. 5C). These data establish the significance of FN in the observed migratory changes in Nudt3KD MCF-7 cells and demonstrate the significance of Nudt3-directed regulation of cell migration through the control of FN mRNA and protein levels. Collectively, Nudt3-depletion elevates RNA and protein expression of ITGB6 and FN, which in turn increases MCF-7 cell motility.
FIGURE 5.
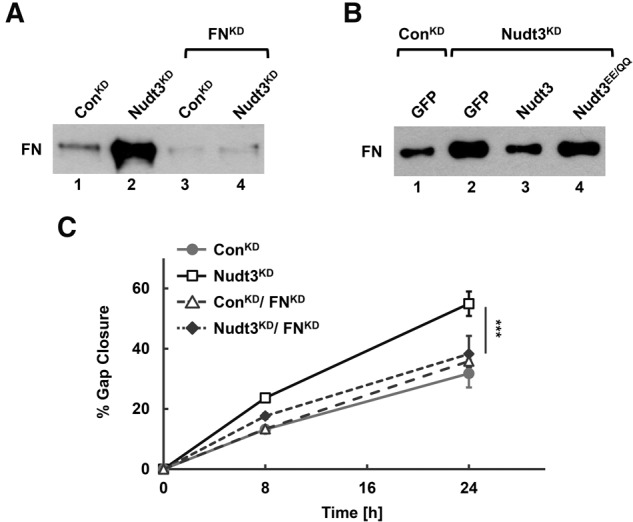
Increased FN secretion in cells with down-regulated Nudt3 is essential for increased MCF-7 cell motility. (A,B) FN protein secreted into the medium was isolated by gelatin–sepharose chromatography from cells grown without FBS on tissue culture plastic. Levels of secreted FN in ConKD and Nudt3KD cells and the same cells with down-regulated FN (A) or complemented with GFP, Nudt3, or Nudt3EE/QQ expression constructs (B) were determined by Western blot analysis. (C) Migration of cells depicted in A were determined by scratch-wound healing assays. At least five different wounds were quantified for each time point. Data are representative of three independent experiments ±SD. P-values are denoted by asterisks. (***) P < 0.001 (Student's t-test).
DISCUSSION
In this report, we demonstrate that the evolutionary conserved Nudt3 hydrolase is an mRNA decapping enzyme that modulates the expression of a subset of mRNAs integral to cell migration. Two of the characterized mRNAs whose steady-state levels were responsive to Nudt3 decapping activity, ITGB6, and LCN2, were shown to be direct targets of Nudt3 decapping function. The stability of both mRNAs was increased upon depletion of Nudt3 and complemented by wild type but not decapping-deficient Nudt3, demonstrating a direct role of Nudt3 in the modulation of ITGB6 and LCN2 mRNAs. The specificity of Nudt3 decapping function was further confirmed by the lack of comparable mRNA alterations in cells diminished for Nudt4 (Figs. 1A, 3E), which lacks decapping activity but shares small molecule dinucleotide hydrolase activity with Nudt3. Moreover, the decapping function of Nudt3 is involved in the modulation of a broader network that controls the expression of genes involved in cell migration, including FN mRNA. Interestingly, regulation of FN mRNA steady-state levels was through an indirect mechanism that was nevertheless dependent on its decapping function. Collectively, our findings support a role for Nudt3 as a decapping enzyme in cells that can directly and indirectly coordinate the steady-state levels of a network of mRNAs involved in cell migration.
In addition to establishing that Nudt3 functions as a decapping enzyme in cells, we further uncovered an important role of Nudt3 in regulating cell motility. Nudt3-depleted MCF-7 cells were characterized by enhanced lamellipodia/filopodia formation as determined by fluorescence staining of actin protein (Fig. 3A) and significantly faster gap closure as demonstrated by in vitro scratch-wound healing assays (Fig. 3B,D). The molecular mechanisms underlying the motility and directed movement of a cell depend on their environment, which comprises the extracellular matrix composition, interaction with other cells, and external stimuli (Ridley 2015). Cell migration involves a number of processes including extension of the leading edge in the direction of movement by protrusion of filopodia or lamellipodia, the formation of adhesion sites between the cell and the substratum, and contraction of the cell to move. Intriguingly, many of the mRNAs regulated by Nudt3 are either cell surface receptors or secreted extracellular proteins involved in cell migration.
A key mediator of Nudt3's influence on cell motility is through its modulation of integrin β6 mRNA stability and resulting protein. αvβ6 expression is highly up-regulated during epithelial remodeling, development, tissue repair, and a variety of neoplastic epithelial cells, including breast cancer (Breuss et al. 1995; Bandyopadhyay and Raghavan 2009). An increase in the normally limiting levels of integrin β6 are expected to increase the abundance of αvβ6 receptor, in turn leading to increased cellular movement. The marked inhibition of cell motility observed upon addition of the αvβ6 receptor-specific blocking antibody in Nudt3KD cells (Fig. 4B) supports a role for this receptor in the Nudt3-directed modulation of cell motility. Additionally, we found that FN, an established ligand for αvβ6, was highly elevated in the media from cells lacking Nudt3 (Fig. 5A,B). Even though stability of FN mRNA was not affected in Nudt3KD cells (Fig. 2), its steady-state transcript (Fig. 1B,D) and protein levels (Fig. 5B) were significantly increased in a Nudt3 decapping-dependent manner. The signaling pathway by which Nudt3 could indirectly modulate FN is not currently known. However, several observations suggest there may be a link between αvβ6 levels and FN expression. Transforming growth factor β (TGF-β) significantly increases FN expression in cells grown on FN and both factors enhance cell migratory behavior (Park and Schwarzbauer 2014). In addition, integrin αvβ6 activates TGF-β (Munger et al. 1999; Annes et al. 2002). Collectively, these observations suggest that the Nudt3-mediated modulation of αvβ6 may indirectly control the expression of FN. We postulate that Nudt3 coordinates MCF-7 cell motility at least in part by increasing the stability of integrin β6 mRNA and in turn αvβ6 receptor and FN protein.
The enhanced migratory phenotype in Nudt3KD cells implies Nudt3 is normally involved in curtailing cell migration. Similar to the ITGB6 and FN mRNAs, LCN2 transcript was also elevated in the absence of Nudt3. Interestingly, proteins encoded by all three Nudt3 target mRNAs have been implicated in increased tumor cell invasion and metastatic potential. The expression of LCN2 is elevated in numerous human epithelial cancers, including breast (Lim et al. 2007), and shown to promote cell migration and invasion (Fougere et al. 2010; Ding et al. 2015). Whether the function of Nudt3 in modulating cell motility is a contributing factor in cellular invasion and metastasis remains to be determined.
In summary, we have identified a third mammalian mRNA decapping enzyme that similar to Dcp2 and Nud16 functions predominantly on a small subset of mRNAs. The lack thus far of a single default mammalian decapping enzyme that nonspecifically targets all mRNAs strongly implies the presence of multiple additional decapping enzymes that preferentially target distinct mRNA subsets to control specific cellular pathways. Whether this is also true in other organisms is not clear. Although Dcp2 appears to be the predominant mRNA decapping enzyme in the yeast S. cerevisiae, the presence of the decapping competent Nudt3 homolog, Ddp1p, suggests there may yet be a population of mRNAs in yeast that are selectively decapped by Ddp1p. Moreover, unlike Nudt16 which is conserved in metazoans, but lacks an obvious ortholog in S. cerevisiae, Caenorhabditis elegans, and Drosophila (Taylor and Peculis 2008), Nudt3 is more highly conserved with orthologs apparent in S. cerevisiae, C. elegans, and Drosophila. Studies are underway to determine whether the additional Nudix proteins that possess in vitro decapping activity in mammals (Nudt-2, -12, -15, -17, -19) and yeast (Ddp1p) (Song et al. 2010) function as decapping enzymes in cells.
MATERIALS AND METHODS
Reagents, cells, and antibodies
Insulin from bovine pancreas, fibronectin from bovine plasma (catalog number F1141) and mitomycin C were purchased from Sigma-Aldrich. The human MCF-7 breast adenocarcinoma was a gift (Fang Liu, Rutgers). pLKO1 plasmids encoding shRNA (MISSION TRC shRNA) against Nudt3 (TRCN0000299497), Nudt4 (TRCN0000296266), FN (TRCN0000293839), and a nonmammalian shRNA control (SHC002V) were purchased from Sigma-Aldrich. Antibodies were obtained from the following sources: BioLegend (LEAF purified IgG2a [κ isotope], catalog number [401504]); EMD Millipore Corporation (αvβ6 [10D5], catalog number MAB2077Z); Thermo Fisher Scientific (Nudt3, catalog number PA5-30435); Santa Cruz Biotechnology, Inc. (integrin β6 [C-19], catalog number sc-6632; GAPDH [0411], catalog number sc-47724); Sigma-Aldrich (fibronectin [clone IST-4], catalog number F0916).
Plasmid constructs
Human Nudt3 open read frame (ORF) was cloned from 293T reverse transcript cDNA by PCR with forward primer 5′-GCTTGGATCCATGATGAAGCTCAAGTCGAACC-3′ and reverse primer 5′-ACTCTCGAGTCATCTGATGCCTGACATCGAGC-3′. BamHI and XhoI restriction sites were added to the forward and reverse primers, respectively, and used for cloning Nudt3 ORF into pcDNA3-Flag vector (Fenger-Gron et al. 2005) to generate pcDNA3-Flag-Nudt3 plasmid. The catalytically inactive Nudix motif mutant pcDNA3-Flag-Nudt3EE/QQ was generated by a one-step mutagenesis PCR approach with forward primer 5′-AAGTCTGTCAGCAGGCTGGAGTAAAAGGGACATTGG-3′ and reverse primer 5′-CTCCAGCCTGCTGACAGACTTCACGAACTGCTGCC-3′. One-step PCR mutagenesis approach was also used for generating Nudt3 shRNA-resistant plasmids pcDNA3-Flag-Nudt3 shRR, and pcDNA3-Flag-Nudt3EE/QQ shRR with forward primer 5′-AGGTAGGTTGGTAGGTATTTTTGAGAACCAGGAGAGGAAGC-3′ and reverse primer 5′-ACCTACCAACCTACCTAATGTCCCTTTTACTCCAGCCTCC-3′ to mutate Nudt3 shRNA target site with no change in Nudt3 protein sequence. pCDH-EF1-Flag-Nudt3shRR and pCDH-EF1-Flag-Nudt3EE/QQ shRR were made by inserting either Flag-Nudt3shRR or Flag- Nudt3EE/QQ shRR flanked by an EcoRI site at the 5′ end and a NotI site at the 3′ end into the pCDH-EF1-MCF-(PGK-GFP-T2A-PURO) (System Bioscience).
Cell culture, transfection, and lentiviral transduction
MCF-7 and HEK293T cells were grown according to ATCC recommendations. Fibronectin coated plates were generated by incubation with a minimal volume of 20 μg/mL fibronectin from bovine plasma (Sigma-Aldrich) in phosphate-buffered saline and air dried for at least 45 min at room temperature. The excess fibronectin was removed and the cells seeded. The lentiviral particles were generated with the pCMV-VSVG and psPAX2 packaging plasmids according to the manufacturer (Thermo Fisher Scientific). Virus-containing supernatants were collected 48 h after transfection and titered. Target cells were infected at a multiplicity of infection (MOI) = 5. Infected cells were selected with 2 μg/mL puromycin (Sigma-Aldrich) 24 h after infection and kept in puromycin for at least 4 d prior to analysis. Nudt3KD cells stably expressing shRNA-resistant wild-type Nudt3 (pCDH-EF1-Flag-Nudt3WTshRR), catalytically inactive Nudt3 mutant (pCDH-EF1-Flag-Nudt3EE/QQshRR), or GFP (empty vector, pCDH-EF1-MCF-[PGK-GFP-T2A-PURO]) were transduced with lentiviral particles generated from respective plasmids.
RNA-seq
RNA-seq libraries were prepared from MCF-7 cells using the Illumina TruSeq RNA Sample Preparation Kit v2 and 2× 100-bp paired-end sequencing was performed by RUCDR Infinite Biologics using the Illumina Hiseq2500 according to Kraushar et al. (2015).
In vitro scratch-wound healing assay
MCF-7 cells were plated in 12-well plates. Confluent cell monolayers were preincubated for 30 min with 0.4 μg/mL mitomycin C (Sigma-Aldrich). A scratch wound was made using a 200 μL pipette tip. Media were replaced with complete medium (10% FBS) containing 0.04 μg/mL mitomycin C or with medium lacking FBS but containing 10 μg/mL anti-αvβ6 integrin (clone 10D5, EMD Millipore Corporation) and 0.04 μg/mL mitomycin C. Wounds were photographed at various time intervals by phase-contrast microscopy with a 20× objective (Zeiss Axio Observer A1). By comparing the images from different time points, we obtained the distance of each scratch closure on the basis of the distance that was measured by ImageJ software. At least 15 individual wound size measurements were averaged at each time point and pooled from three independent experiments. The data are presented as a percentage of gap closure over time.
Western blotting and immunofluorescence
Cells were lysed in phosphate-buffered saline containing 0.5% IGEPAL CA-630 (Sigma-Aldrich) and protease inhibitors (Roche Applied Science), and sonicated. Equal protein amounts of the different samples were separated by 15% or 6% SDS-PAGE and transferred to nitrocellulose membranes (Bio-Rad). The blots were incubated with primary antibodies in phosphate-buffered saline buffer supplemented with 5% BSA (Sigma-Aldrich), then with secondary antibodies conjugated to horseradish peroxidase. Proteins were detected using the ECL Western blotting substrate (Thermo Fisher Scientific). MCF-7 cells, cultured on glass coverslips in six-well plates to 70% confluency, were fixed with 3.7% formaldehyde solution in PBS for 10 min at room temperature and permeabilized with acetone at −20°C for 3 min. The coverslips were blocked with 3% bovine serum albumin (BSA) at room temperature and cells were stained with Alexa Fluor 488 conjugated phalloidin (Invitrogen) at room temperature for 30 min according to the manufacturer's suggested dilution. Nuclei were stained with DAPI. All immunofluorescence images were captured by fluorescent microscopy with a 40× objective (Zeiss Axiovert 100M Microscope) and processed with Adobe Photoshop software.
Measurements of fibronectin secretion
Semi-confluent MCF-7 cells plated in 35-mm dishes were incubated overnight in 1.5 mL DMEM with 1% FBS. To measure fibronectin synthesis, 1 mL aliquots of culture media were incubated with 100 μL gelatin Sepharose 4B (GE Healthcare) beads overnight at 4°C. The beads were washed three times in PBS and bound fibronectin was eluted from the beads into 100 μL Laemmli sample buffer by boiling for 5 min. The samples were then separated on 6% SDS-polyacrylamide gels and fibronectin detected by Western blotting.
RNA isolation, reverse-transcription, and real-time qRT-PCR
Total cellular RNA was harvested with TRIzol Reagent (Thermo Fisher Scientific) and treated with RNase-free DNase (Promega) according to the manufacturers’ protocols. Reverse transcription was performed on 2 μg of RNA in 20-μL reaction mixtures with M-MLV reverse transcriptase, random hexamers, and oligo(dT) (Promega) according to the manufacturer's instructions. qRT-PCR was performed with the primers listed in Table 1 and carried out on Rotor-Gene 3000 (Corbett Research) with iTaq SYBR Green Supermix (Bio-Rad Laboratories). Relative mRNA levels were normalized to 18S rRNA and calculated as described in User Bulletin No. 2 for the ABI Prism 7700 Sequence Detection System.
TABLE 1.
Primers list

DATA DEPOSITION
Sequencing data have been deposited in the Gene Expression Omnibus (GEO) database (accession number GSE75146).
ACKNOWLEDGMENTS
We thank F. Matsumura (Rutgers) and members of the Kiledjian laboratory for helpful discussion. This work was supported by National Institutes of Health (NIH) grant GM067005 to M.K.
Footnotes
Article published online ahead of print. Article and publication date are at http://www.rnajournal.org/cgi/doi/10.1261/rna.055699.115.
REFERENCES
- Annes JP, Rifkin DB, Munger JS. 2002. The integrin αVβ6 binds and activates latent TGFβ3. FEBS Lett 511: 65–68. [DOI] [PubMed] [Google Scholar]
- Bandyopadhyay A, Raghavan S. 2009. Defining the role of integrin αvβ6 in cancer. Curr Drug Targets 10: 645–652. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bessman MJ, Frick DN, O'Handley SF. 1996. The MutT proteins or “Nudix” hydrolases, a family of versatile, widely distributed, “housecleaning” enzymes. J Biol Chem 271: 25059–25062. [DOI] [PubMed] [Google Scholar]
- Breuss JM, Gallo J, DeLisser HM, Klimanskaya IV, Folkesson HG, Pittet JF, Nishimura SL, Aldape K, Landers DV, Carpenter W, et al. 1995. Expression of the β6 integrin subunit in development, neoplasia and tissue repair suggests a role in epithelial remodeling. J Cell Sci 108(Pt 6): 2241–2251. [DOI] [PubMed] [Google Scholar]
- Chen CY, Shyu AB. 2011. Mechanisms of deadenylation-dependent decay. Wiley Interdiscip Rev RNA 2: 167–183. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Coller J, Parker R. 2004. Eukaryotic mRNA Decapping. Annu Rev Biochem 73: 861–890. [DOI] [PubMed] [Google Scholar]
- Ding G, Fang J, Tong S, Qu L, Jiang H, Ding Q, Liu J. 2015. Over-expression of lipocalin 2 promotes cell migration and invasion through activating ERK signaling to increase SLUG expression in prostate cancer. Prostate 75: 957–968. [DOI] [PubMed] [Google Scholar]
- Fenger-Gron M, Fillman C, Norrild B, Lykke-Andersen J. 2005. Multiple processing body factors and the ARE binding protein TTP activate mRNA decapping. Mol Cell 20: 905–915. [DOI] [PubMed] [Google Scholar]
- Fougere M, Gaudineau B, Barbier J, Guaddachi F, Feugeas JP, Auboeuf D, Jauliac S. 2010. NFAT3 transcription factor inhibits breast cancer cell motility by targeting the Lipocalin 2 gene. Oncogene 29: 2292–2301. [DOI] [PubMed] [Google Scholar]
- Furuichi Y, LaFiandra A, Shatkin AJ. 1977. 5′-Terminal structure and mRNA stability. Nature 266: 235–239. [DOI] [PubMed] [Google Scholar]
- Gallie DR. 1991. The cap and poly(A) tail function synergistically to regulate mRNA translational efficiency. Genes Dev 5: 2108–2116. [DOI] [PubMed] [Google Scholar]
- Hofker M, Wijmenga C. 2009. A supersized list of obesity genes. Nat Genet 41: 139–140. [DOI] [PubMed] [Google Scholar]
- Hsu CL, Stevens A. 1993. Yeast cells lacking 5′→3′ exoribonuclease 1 contain mRNA species that are poly(A) deficient and partially lack the 5′ cap structure. Mol Cell Biol 13: 4826–4835. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Huang X, Wu J, Spong S, Sheppard D. 1998. The integrin αvβ6 is critical for keratinocyte migration on both its known ligand, fibronectin, and on vitronectin. J Cell Sci 111(Pt 15): 2189–2195. [DOI] [PubMed] [Google Scholar]
- Humphries JD, Byron A, Humphries MJ. 2006. Integrin ligands at a glance. J Cell Sci 119: 3901–3903. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kraushar ML, Viljetic B, Wijeratne HR, Thompson K, Jiao X, Pike JW, Medvedeva V, Groszer M, Kiledjian M, Hart RP, et al. 2015. Thalamic WNT3 secretion spatiotemporally regulates the neocortical ribosome signature and mRNA translation to specify neocortical cell subtypes. J Neurosci 35: 10911–10926. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Li Y, Song MG, Kiledjian M. 2008. Transcript-specific decapping and regulated stability by the human Dcp2 decapping protein. Mol Cell Biol 28: 939–948. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Li Y, Song MG, Kiledjian M. 2011. Differential utilization of decapping enzymes in mammalian mRNA decay pathways. RNA 17: 419–428. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Li Y, Dai J, Song M, Fitzgerald-Bocarsly P, Kiledjian M. 2012. Dcp2 decapping protein modulates mRNA stability of the critical interferon regulatory factor (IRF) IRF-7. Mol Cell Biol 32: 1164–1172. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Liang CC, Park AY, Guan JL. 2007. In vitro scratch assay: a convenient and inexpensive method for analysis of cell migration in vitro. Nat Protoc 2: 329–333. [DOI] [PubMed] [Google Scholar]
- Lim R, Ahmed N, Borregaard N, Riley C, Wafai R, Thompson EW, Quinn MA, Rice GE. 2007. Neutrophil gelatinase-associated lipocalin (NGAL) an early-screening biomarker for ovarian cancer: NGAL is associated with epidermal growth factor-induced epithelio-mesenchymal transition. Int J Cancer 120: 2426–2434. [DOI] [PubMed] [Google Scholar]
- Lykke-Andersen J. 2002. Identification of a human decapping complex associated with hUpf proteins in nonsense-mediated decay. Mol Cell Biol 22: 8114–8121. [DOI] [PMC free article] [PubMed] [Google Scholar]
- McLennan AG. 2006. The Nudix hydrolase superfamily. Cell Mol Life Sci 63: 123–143. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Muhlrad D, Decker CJ, Parker R. 1995. Turnover mechanisms of the stable yeast PGK1 mRNA. Mol Cell Biol 15: 2145–2156. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Munger JS, Huang X, Kawakatsu H, Griffiths MJ, Dalton SL, Wu J, Pittet JF, Kaminski N, Garat C, Matthay MA, et al. 1999. The integrin αvβ6 binds and activates latent TGF β 1: a mechanism for regulating pulmonary inflammation and fibrosis. Cell 96: 319–328. [DOI] [PubMed] [Google Scholar]
- Park J, Schwarzbauer JE. 2014. Mammary epithelial cell interactions with fibronectin stimulate epithelial-mesenchymal transition. Oncogene 33: 1649–1657. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pereira IT, Ramos EA, Costa ET, Camargo AA, Manica GC, Klassen LM, Chequin A, Braun-Prado K, Pedrosa Fde O, Souza EM, et al. 2014. Fibronectin affects transient MMP2 gene expression through DNA demethylation changes in non-invasive breast cancer cell lines. PLoS One 9: e105806. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Preiss T, Hentze MW. 1998. Dual function of the messenger RNA cap structure in poly(A)-tail-promoted translation in yeast. Nature 392: 516–520. [DOI] [PubMed] [Google Scholar]
- Ridgway PF, Ziprin P, Peck DH, Darzi AW. 2005. Hypoxia increases reepithelialization via an αvβ6-dependent pathway. Wound Repair Regen 13: 158–164. [DOI] [PubMed] [Google Scholar]
- Ridley AJ. 2015. Rho GTPase signalling in cell migration. Curr Opin Cell Biol 36: 103–112. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shears SB. 2009. Diphosphoinositol polyphosphates: metabolic messengers? Mol Pharmacol 76: 236–252. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Song MG, Li Y, Kiledjian M. 2010. Multiple mRNA decapping enzymes in mammalian cells. Mol Cell 40: 423–432. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Song MG, Bail S, Kiledjian M. 2013. Multiple Nudix family proteins possess mRNA decapping activity. RNA 19: 390–399. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Taylor MJ, Peculis BA. 2008. Evolutionary conservation supports ancient origin for Nudt16, a nuclear-localized, RNA-binding, RNA-decapping enzyme. Nucleic Acids Res 36: 6021–6034. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Trapnell C, Hendrickson DG, Sauvageau M, Goff L, Rinn JL, Pachter L. 2013. Differential analysis of gene regulation at transcript resolution with RNA-seq. Nat Biotechnol 31: 46–53. [DOI] [PMC free article] [PubMed] [Google Scholar]
- van Dijk E, Cougot N, Meyer S, Babajko S, Wahle E, Seraphin B. 2002. Human Dcp2: a catalytically active mRNA decapping enzyme located in specific cytoplasmic structures. EMBO J 21: 6915–6924. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang Z, Jiao X, Carr-Schmid A, Kiledjian M. 2002. The hDcp2 protein is a mammalian mRNA decapping enzyme. Proc Natl Acad Sci 99: 12663–12668. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Weinacker A, Chen A, Agrez M, Cone RI, Nishimura S, Wayner E, Pytela R, Sheppard D. 1994. Role of the integrin αvβ6 in cell attachment to fibronectin. Heterologous expression of intact and secreted forms of the receptor. J Biol Chem 269: 6940–6948. [PubMed] [Google Scholar]
- Williams MJ, Eriksson A, Shaik M, Voisin S, Yamskova O, Paulsson J, Thombare K, Fredriksson R, Schioth HB. 2015. The obesity-linked gene Nudt3 Drosophila homolog Aps is associated with insulin signaling. Mol Endocrinol 29: 1303–1319. [DOI] [PMC free article] [PubMed] [Google Scholar]