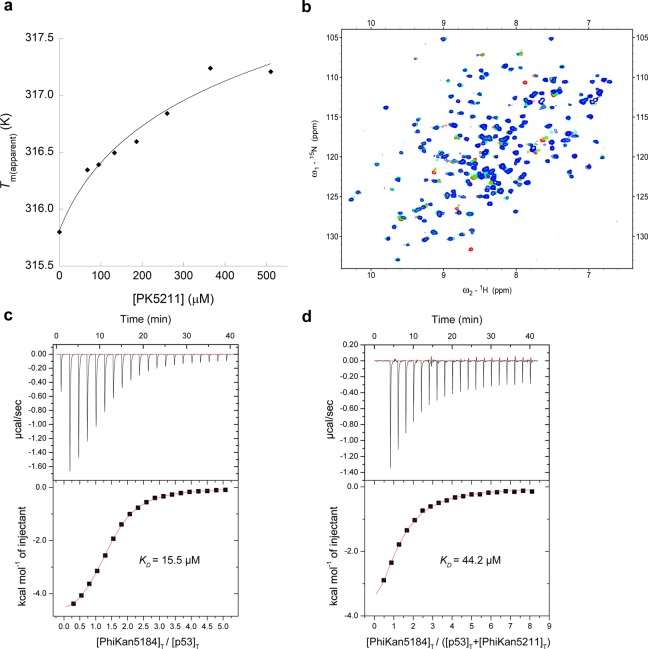
Figure 2.
Biophysical characterization of PK5211 (3) binding to p53-Y220C DNA-binding domain. (a) Differential scanning fluorimetry (DSF) shows concentration-dependent thermal stabilization of the mutant protein. (b) 1H/15N-HSQC NMR shows that 3 perturbs and quenches specific residue signals upon binding, consistent with its binding to the mutation-induced surface crevice. (c, d) Competition ITC experiments: PK5174 (1) binds to p53-Y220C with KD = 15.5 μM; its binding affinity is shifted to KD = 44.2 μM upon addition of 500 μM of compound 3. The resulting affinity of 3 for p53-Y220C was then calculated as KD = 271 μM.24