Figure 1.
High-Resolution Quantitative MS Reveals Transient HCV Entry Factor Interactions
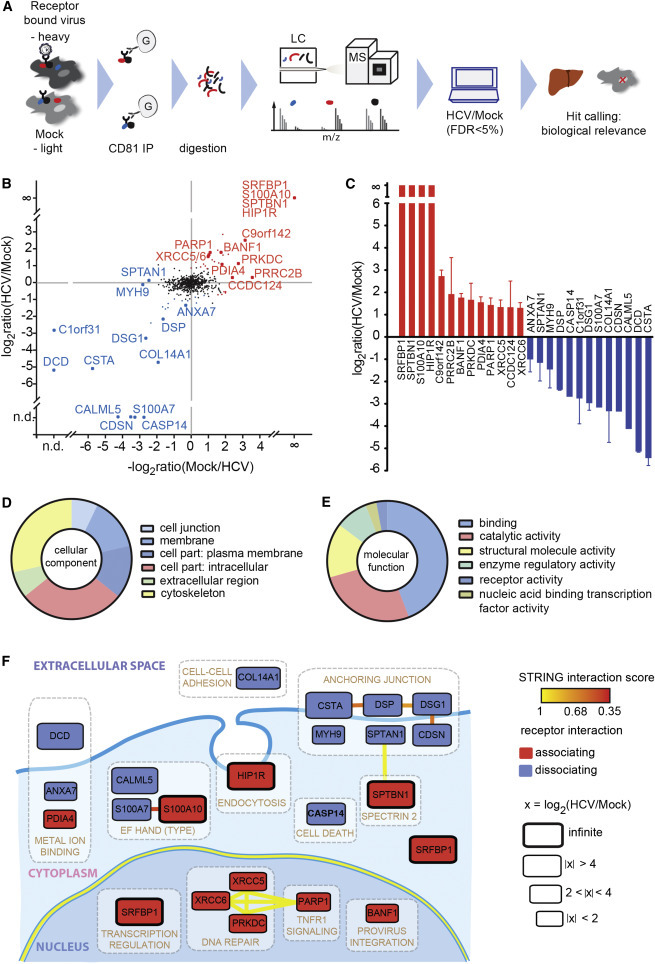
(A) Outline of the virus entry interaction proteomics procedure.
(B) CD81 interactome upon HCV exposure. Depicted are the mean log2 SILAC ratios of CD81-interacting proteins in HCV versus mock-treated samples from forward (y axis) and reverse experiments (x axis). Reverse label ratios are inverted, so that a positive correlation indicates reproducible interaction upon label swap. Significant (FDR < 5%) outliers are colored in red (CD81-associating proteins) and blue (CD81-dissociating proteins). Infinite ratio, interaction partners exclusively found in the presence of HCV. n.d., not quantified in either forward or reverse experiment.
(C) SILAC log2 ratios for each of the 13 CD81-associating and 13 CD81-dissociating factors. Shown are means ± SEM of four biological replicates with inverted reverse label ratios.
(D and E) Enrichment of Gene Ontology cellular component (GOCC) and molecular function (GOMF) annotations.
(F) Functional map of host factors transiently interacting with the HCV receptor CD81 during virus entry. Functional clusters (white boxes) and previously reported interactions (bold lines) of the here identified transient CD81-binding partners are depicted. We assigned individual proteins to the highest scoring DAVID cluster. Yellow lines between genes of different clusters indicate high-confidence (>0.9) STRING interactions. Within a functional annotation cluster, also lower confidence (>0.35) STRING interactions are shown. Proteins are placed in their predominant cellular location; SRFBP1 is shown twice as it localizes to nucleus and cytoplasm. The box size indicates the degree of CD81 association or dissociation upon HCV binding. Associating factors (red) and dissociating factors (blue) are shown. See also Figure S1 and Tables S1 and S2–S4.
