Figure 6.
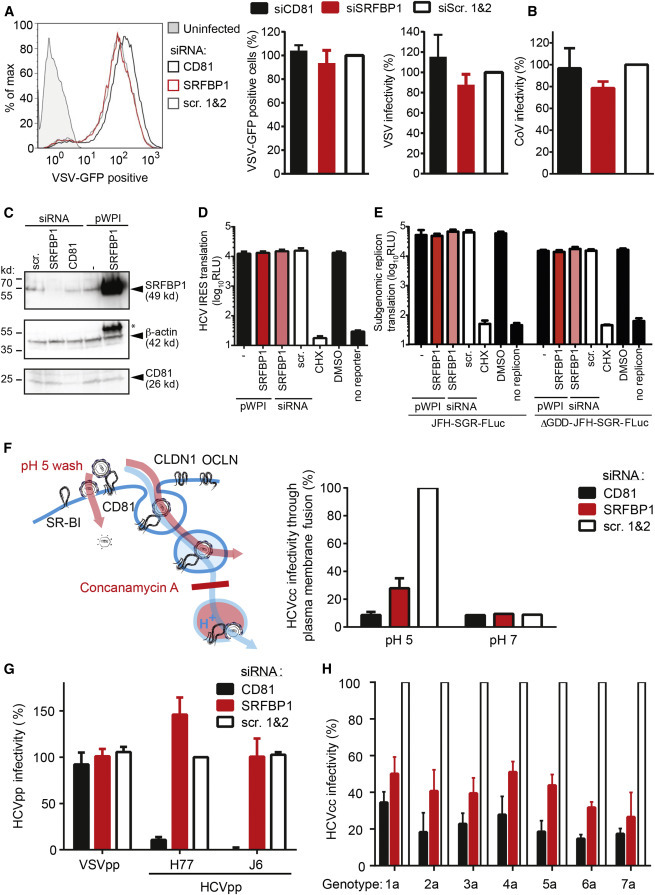
SRFBP1 Is a Pan-genotypic and HCV-Specific Host Entry Factor
(A) SRFBP1 is dispensable for VSV infection. SRFBP1-silenced Huh-7.5 cells were infected with VSV∗MQ (MOI 0.1) and analyzed for GFP expression by flow cytometry 20 hpi. Histogram is representative of biological triplicates (left panel). Quantification of VSV∗MQ infectivity 20 hpi in SRFBP1-silenced cells is determined as percentage of GFP-positive cells (middle panel) or by normalization of the mean fluorescence intensity (MFI) of VSV-infected SRFBP1- or CD81-silenced cells to MFI of scrambled siRNA-transfected cells (right panel).
(B) SRFBP1 is dispensable for coronavirus infection. SRFBP1-silenced cells were infected with HCoV229E-luc (MOI 0.1) and RLuc activity in cell lysates measured 24 hpi. Infectivity relative to a scrambled siRNA control is shown.
(C) Immunoblot analysis of SRFBP1 and CD81 48 hp siRNA transfection. Huh-7.5 cells were transfected with siRNA or transduced with the indicated pWPI expression construct as in (A)–(H), 48 hr later lysed, and analyzed by immunoblot. Actin served as loading control. ∗, residual SRFBP1 signal.
(D) Bicistronic translational reporter assay with HCV IRES-driven RLuc and cap-dependent FLuc (see also Figures S6A and S6B). SRFBP1 silencing and overexpression was performed as in (C), and 48 hr later, cells were transfected with translational reporter RNA. Eight hours after reporter transfection, luciferase activity in lysates was monitored.
(E) Early replication reporter assay using a subgenomic HCV genome expressing FLuc. SRFBP1 silencing and overexpression was performed as in (C), and 48 hr later, JFH-SGR-FLuc RNA was transfected into cells; cells lysed after 8 hr; and luciferase activity monitored. A polymerase mutant JFH-SGR-FLuc replicon (ΔGDD) was used to assess translation of HCV genomes independent of de novo replication. See also Figures S6C and S6D for additional controls.
(F) SRFBP1 is required in a plasma membrane fusion assay of HCV infection. Huh-7.5 cells silenced for SRFBP1 were pretreated with concanamycin A (5 nM; 1 hr) to block vacuolar type H+-ATPases, incubated with HCV (JcR2A) for 2 hr at 4°C in the presence of concanamycin A, shifted to 37°C, and washed with a pH 5 or pH 7 buffer for 5 min. After incubation with concanamycin A for 4 hr, medium was changed and endosomal acidification independent infectivity measured at 48 hpi. See also Figures S6E and S6F for additional controls.
(G) Lentiviral pseudotypes infect Huh-7.5 cells independently of SRFBP1. Cells in which SRFBP1 had been silenced (48 hr) were infected with HIV-1 pseudotypes encoding FLuc and displaying glycoproteins from HCV genotype 1 (H77), HCV genotype 2 (J6), VSV, or no glycoprotein. At 72 hpi, cells were lysed and FLuc activity measured. Infectivity was calculated by subtraction of background read for glycoprotein-free particles and relative to VSVG particles.
(H) Silencing of SRFBP1 reduces infectivity of chimeric HCV viruses with glycoproteins from all seven genotypes. Huh-7.5 FLuc cells were subjected to siRNA-mediated silencing followed by infection with intergenotypic HCV chimeras (MOI 0.1) expressing RLuc. Forty-eight hours post-infection, infectivity was determined by RLuc activity measurement. Cells treated with CD81 targeting or scrambled siRNAs served as controls. SRFBP1-targeting siRNA 394 was used in all experiments. Data from three to five biological replicates are displayed as mean + SD. See also Figure S6.
