Abstract
Biomedical ontologies contain errors. Crowdsourcing, defined as taking a job traditionally performed by a designated agent and outsourcing it to an undefined large group of people, provides scalable access to humans. Therefore, the crowd has the potential overcome the limited accuracy and scalability found in current ontology quality assurance approaches. Crowd-based methods have identified errors in SNOMED CT, a large, clinical ontology, with an accuracy similar to that of experts, suggesting that crowdsourcing is indeed a feasible approach for identifying ontology errors. This work uses that same crowd-based methodology, as well as a panel of experts, to verify a subset of the Gene Ontology (200 relationships). Experts identified 16 errors, generally in relationships referencing acids and metals. The crowd performed poorly in identifying those errors, with an area under the receiver operating characteristic curve ranging from 0.44 to 0.73, depending on the methods configuration. However, when the crowd verified what experts considered to be easy relationships with useful definitions, they performed reasonably well. Notably, there are significantly fewer Google search results for Gene Ontology concepts than SNOMED CT concepts. This disparity may account for the difference in performance – fewer search results indicate a more difficult task for the worker. The number of Internet search results could serve as a method to assess which tasks are appropriate for the crowd. These results suggest that the crowd fits better as an expert assistant, helping experts with their verification by completing the easy tasks and allowing experts to focus on the difficult tasks, rather than an expert replacement.
Keywords: crowdsourcing, ontology engineering, Gene Ontology
Graphical Abstract
1. Introduction
Ontologies enable researchers to specify, in a computational fashion, the entities that exist in the world, their properties, and their relationships to other entities. For instance, a researcher might encode in an ontology the kinds of cellular components that exist, such as a nucleus or ribosome. By leveraging such an ontology, a computer can recognize that a nucleus and a ribosome are, in fact, both a kind of cellular component and use that relationship when aggregating data. Further, ontologies allow everyone to “speak the same language” by creating a shared set of terms with clearly defined meanings. This property enables disparate parties to share data and to integrate them readily. For example, when two data sources contain different information about cellular components (one focused on nuclei and the other on ribosomes) and use the same ontology to describe that information, a researcher is able to combine them with relative ease. These powerful properties enable ontologies to facilitate data integration, search, decision support, and data annotation [1]. Today, ontologies are ubiquitous. Indeed, the Google Knowledge Graph contains an ontology that supports an advanced understanding of entities on the Internet. With the Knowledge Graph’s ontology, Google provides additional information about an entity – a search for a movie also provides its star actors, director, budget, and so on [2]. Ontologies are latent in many of the technologies we encounter today. Given the important of ontologies, it is essential to ensure users are able to build and maintain them with minimal errors. In this work, we consider applying crowdsourcing to the task of ontology quality assurance – a task that is particularly challenging for biomedical ontologies.
Biomedicine relies heavily on ontologies. In the clinic, they support electronic health records with tasks such as computerized physician order entry, alerting, and decision support [3]. In the life sciences, ontologies help combat the data deluge, giving researchers a tool to describe the intricate complexities of biomedicine and use that encoded knowledge to organize, annotate, and sift through data [4–7]. One of the most well known biomedical ontologies is the Gene Ontology [8]. By describing, in a computational fahsion, experimental data and published literature with Gene Ontology (GO) terms, researchers are able to integrate results that are described with the same terms and gain insight about cellular components, biological processes, and molecular functions involved with a gene set of interest. One common use of these annotations and terms is GO enrichment analysis, wherein sets of differentially expressed genes are related, via a statistical over-representation analysis, to terms in GO [9]. These returned terms assist a researcher in developing hypotheses about the underlying biological phenomena that differentiates cases and controls. Of note, when one works with microarray data, GO enrichment analyses are standard practice. Such studies are pervasive in the literature.
The Gene Ontology its application is just one of the many examples the rapid increase in ontology use. Demonstrating this trend, The National Center for Biomedical Ontology provides a repository, called the Bioportal, of over 450 ontologies ranging from brain anatomy to medical procedures [10]. These ontologies vary in size from hundreds of concepts to tens of thousands concepts and contain even more relationships between those concepts. However, as the size and complexity of ontologies continue to grow, so too does the difficulty of their development and maintenance. It becomes difficult for any single engineer to grasp the entirety of the ontology.
As a consequence of the difficulty of ontology development and maintenance, ontologies, not surprisingly, contain errors. Rector[11], Ceusters[12, 13], Mortensen[14], and others have all identified systematic issues in SNOMED CT, an ontology intended to describe clinical encounters, and the National Cancer Institute Thesaurus, a clinical ontology focused on cancer. SNOMED CT contained doman-specific errors such as Short Sleeper SubClassOf Brain Disorder (brain disorders are not the sole cause of short sleep) and Diabetes SubClassOf Disorder of the Abdomen (diabetes is not a disorder of the abdominal cavity but rather of the endocrine system). In this work, we refer to techniques that identify such errors as “ontology verification”. Speaking to the frequency of these errors, there have been entire journal special issues dedicated to ontology quality verification methods [15]. Unfortunately, these methods are limited in their ability to catch domain-specific errors. For instance, a common class of computational ontology evaluation methods is metrics-based. In these methods, metrics are calculated about various characteristics of an ontology, such as its structure (e.g., average number of children), its syntax (e.g., number of syntax errors), its content (e.g., number of definitions) or adherence to best practices (e.g., using fully defined concepts) [16–22]. These metrics serve as a proxy for ontology quality. However, quality alone does not point to specific errors, limiting these methods in their ability to find errors such as those highlighted above (i.e., domain-specific errors). As a result, the currently accepted approach for identifying ontology errors is expert review. Only domain experts can interpret the symbols in an ontology and determine whether they reflect their understanding of the domain. However, the use of experts is very expensive. Experts cannot verify the large ontologies now found in biomedicine simply by inspection. In short, there is a fundamental trade-off between scalability (computational) and accuracy (expert) in current ontology verification methods.
Crowdsourcing, the practice of taking work traditionally done by one person and outsourcing it to online, anonymous crowds [23], is one approach to overcoming the limitations existing ontology quality assurance methods. Researchers have shown that crowdsourcing can solve certain intuitive, human-level intelligence tasks more accurately than computers. For example, crowds of online workers might annotate an image with properties such as whether it contains a ball or a cat. Performing this task computationally remains a challenge, but humans can complete it easily. As crowdsourcing has grown, online platforms have emerged that provide users (i.e., requesters) with access to crowds (i.e., workers). The most common form of crowdsourcing on these platforms is micro-tasking. Here, many workers complete small, short tasks (requiring only minutes) for small rewards, including monetary compensation [23]. With this model, large tasks are completed quickly by large crowds that scale dynamically. Crowdsourcing is a complement to many computational techniques.
Researchers have begun using crowdsourcing extensively [24–26]. One challenge that remains in crowdsourcing research is understanding how the crowd can contribute to solving expert-level, knowledge-intensive tasks. In the biomedical realm, for one such expert-level domain, MacLean and Heer developed a crowd-based methodology to extract medical entities from patient-authored text[27]. They used crowd workers to find and to label terms. They then used these labels as a training set for a statistical classifier. This classifier then identified relevant medical terms written by patients in online forums. This system was able to identify medical terms with significantly higher accuracy in comparison to common automated medical extraction methods and thus showed that the crowd can work reliably on certain medical topics.
The use of crowdsourcing in ontology engineering, a knowledge-intensive task, is still nascent. There has been beginning investigation into micro-task based ontology mapping and gaming-based ontology tagging [28–30]. The success of this work suggests that crowdsourcing is a candidate to solve various ontology engineering tasks. Building on these efforts, in our previous work, we have developed, refined, and applied methods to perform ontology verification with the crowd [31, 32]. At a high level, the method asks crowd workers to read sentences reflecting natural language representations of relationships in an ontology and to decide whether a sentence is True or False based on their knowledge and provided definitions. We have already applied successfully this method to verify a sample of SNOMED CT, finding a number of errors (More detail in Section 2)[14].
In this work, we applied the same crowd-based verification methodology to another ontology, the Gene Ontology. We investigated how the crowd performed in various configurations and how their performance varied with task difficulty and the quality of concept definitions. Further, we developed a strategy to predict a task’s difficulty based on Google search results. In doing so, we make the following contributions:
We replicated previous work on crowdsourced ontology verification.
We compared and contrast our results on verifying GO with those of SNOMED CT.
We identified the important factors required for successfully using crowd-sourcing for ontology verification.
We described a system for a hybrid between crowd-based and expert-based ontology verification (i.e. “group-sourcing”).
2. SNOMED CT verification study summary
The current work is based on on our previous work[14]. Here, we summarize the results of that work. Note that the methodology is the same for both studies, and therefore Section 3 details the methodology itself.
We created a hierarchical verification crowdsourcing task template. Figure 1 shows an instantiation of the template asking workers to determine whether “Microcephalus is a kind of Disorder of brain”. We applied this template to SNOMED CT, an ontology that specifies a set of concepts, terms, and relationships relevant for clinical documentation. These concepts range from drugs and procedures to diagnoses and human anatomy. We focused on 200 relationships from SNOMED CT (January 2013 version) that were widely-used across US hospitals and were likely to contain errors. We asked 25 workers to complete each of the 200 tasks, compensating them $0.02 per task and then aggregated their responses into a final decision [33]. Experts completed the same verification task in parallel via an online survey to provide a gold standard of comparison.
Figure 1. Example SNOMED CT verification task.
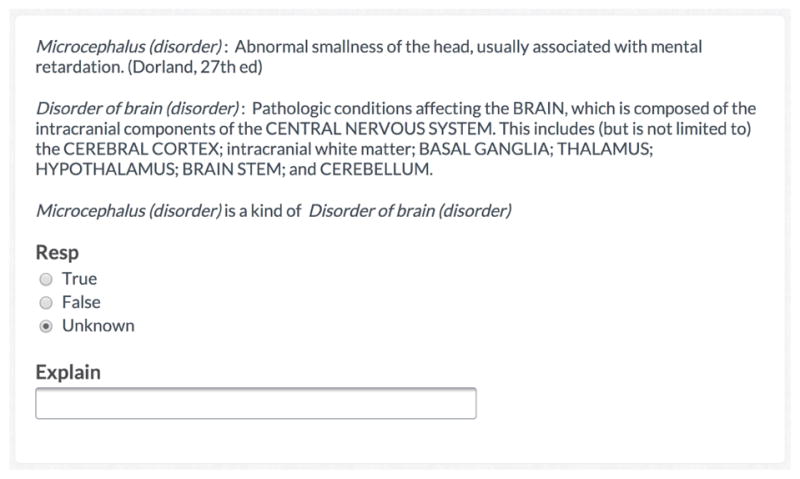
An instantiation of the crowdsourcing task template with which crowd workers verified SNOMED CT
The crowd identified 39 errors (20% of the 200 relationships we verified), was indistinguishable from any single expert by inter-rater agreement (expert vs. expert kappa: 0.58; crowd vs. expert kappa: 0.59), and performed on par with any single expert against the gold standard, with a mean area under the receiver operating characteristic curve of 0.83. In addition, the crowd cost one quarter that of experts ($0.50/relationship vs. $2.00/relationship). These results suggested that the crowd can indeed approximate expert capability on SNOMED CT. Further, they suggested the crowd is best suited for verification situations with limited budgets, a lack of experts, or very large biomedical ontologies. With these promising results, we moved to test the methodology on the Gene Ontology. Unlike SNOMED CT, which is intended for detailed descriptions on a breadth of clinical encounters, the Gene Ontology is used primarily to annotate documents and describes a more specialized field.
3. Methods
In the current work, we investigated the verification of the Gene Ontology. To show comparability, the methods in this work closely mirror those of our previous work on SNOMED CT [14]. We first extracted a manageable, logically complex subset from GO and created expert-based consensus standard of errors for that subset. We then used the crowd-based method (in various configurations) to verify the extracted subset of GO. We analyzed the crowd results using standard statistical comparisons, focusing on the impact that various method configurations and task factors had on the method’s performance.
Gene Ontology
In this work, we evaluated a subset of the GO Plus version of GO from April 2014. This version adds “cross ontology relationships (axioms) and imports additional required ontologies” resulting in a more semantically rich and complex ontology1. To select a manageable portion of GO Plus for verification (200 relationships), we used a modified version of the filtering criteria we developed previously [34]. The goal of these criteria is to find complex relationships in the ontology that are likely more error-prone because experts do not create them directly. In practice, we encoded the following filtering criteria for relationships using the OWL API, a software tool for working with ontologies in a programmatic fashion [35]:
Non-Trivial A relationship that is not explicitly stated but is instead logically entailed by the interaction of two or more axioms. This step removes those relationships that were explicitly specified by a human curator.
Direct A hierarchical relationship between two concepts where no concept exists in the inferred hierarchy between those two concepts. This step removes relationships (i.e., subclass axioms to ancestors of the immediate parent) that describe a very simple hierarchical relationship. Note that such relationships are always generated by classifiers but ontology visualization tools may not always show them.
Complete text definitions Both concepts in the relationship have a textual definition in GO. In previous work, we found that definitions are key to successful crowdsourcing [32].
Applying these criteria selects 329 relationships from GO Plus. We then randomly selected 200 relationships from that resulting set to have the same number of relations as verified in the SNOMED CT study [14]. Figure 2 summarizes the selection process.
Figure 2. Filtering GO relationships to a manageable subset.

To select a manageable, complex subset of Gene Ontology relationships, we applied a filtering process that selects relationships that are (1) not explicitly specified by the ontology developers but instead logically entailed and (2) contained concepts with explicit text definitions.
Expert Verification of GO
To measure the ability of the crowd-based method, we first developed an expert-based consensus standard against which to compare it. The methodology in this work is the same as prior work and a more extensive description of the method is available there [34]. Five authors with expertise in biology, cell biology, biochemistry, ontology and bioinformatics (NT, JJH, HFM, KVA, and MD) verified the 200 selected relations in GO following the same format devised in our prior work. This verification process had two stages: an initial verification survey followed by a survey designed to resolve inconsistencies in expert responses. In the first stage, an online survey showed each expert, for each relationship, two concepts, their definition, and a natural language representation of the GO relationship. With that information, and their background knowledge, each expert indicated whether they believed the relationship to be correct or incorrect. Figure 3 depicts one of the 200 survey questions each expert completed.
Figure 3. Example expert verification survey.
To create a consensus standard of errors in the GO relationship subset, we asked experts, the gold-standard method for verification, to complete an online survey in which they assessed the correctness of the selected relationships.
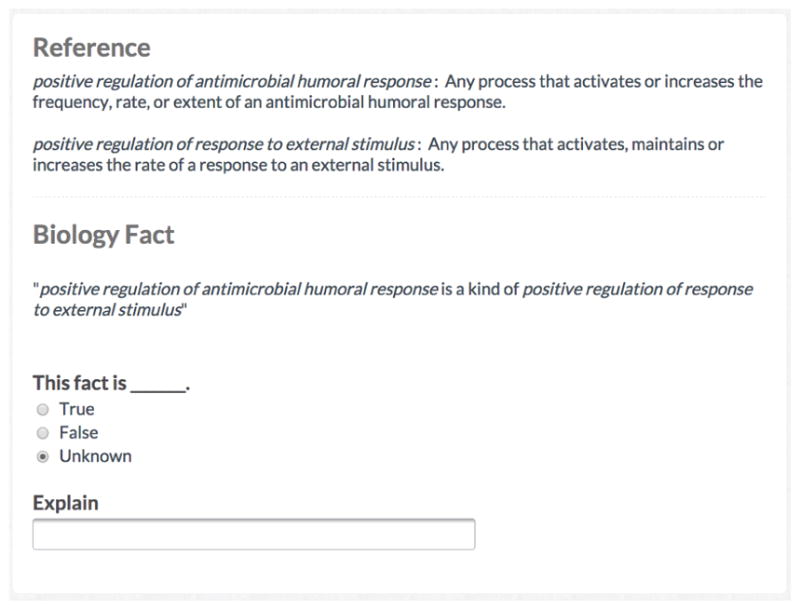
In the second stage, after all experts completed the survey, we identified relationships about which they disagreed and asked them to reach consensus via a second survey. This survey followed the Delphi method [36], wherein experts viewed an anonymous set of responses and comments from other experts and then reconsidered their initial response in light of this new information. Figure 4 shows an example of this survey. With the responses collected, we created an expert-based consensus standard of the error status of the 200 GO relationships by using a super-majority vote (i.e., 4 out of 5 experts). We excluded from the standard those relationships on which experts could not achieve super-majority agreement.
Figure 4. Example expert Delphi survey.
Once experts completed the initial survey, we asked them to reach consensus on areas where all five did not agree. Experts completed another survey where they read the anonymous responses and comments that all experts made about the relationships and then updated their response considering this new information. Formally, this process is known as the Delphi method. With consensus obtained, we then had a reference standard of errors against which to compare the crowd.
Crowd-Based Verification of GO
We asked the crowd to verify the 200 GO Plus relationships using the methodology developed previously [14]. Following that method, we thus submitted tasks to an online crowdsourcing platform, presenting workers with a verification task similar to that which the experts completed. Specifically, our task asked workers to read a natural language representation of a GO relationship and to determine whether that sentence is True or False based on their knowledge and a set of provided definitions. Figure 5 shows a screenshot of such task. Workers were compensated for the completion of each verification task.
Figure 5. Example crowd task on CrowdFlower.
In parallel to the expert verification, we submitted the same ontology verification task to two online platforms, CrowdFlower and Mechanical Turk, in various compensation and quality filtering configurations.
After we received 25 worker responses for each verification task, we combined them using the unsupervised aggregation technique developed by Simpson and colleagues [33]. The intuition behind this method is that it attempts to determine the probability that each relationship is “correct” by estimating worker ability and task difficulty. In short, the methodology treats worker responses as samples that are drawn from a multinomial distribution that is parameterized by Dirichlet distributions designed to model worker ability and task difficulty. A variational Bayes approach (similar to Expectation Maximization) is used to arrive at approximate parameters that describe these Direchlet distributions and that predict the observed data. As a side effect, the Dirichlet distributions also predict worker specificity and sensitivity. Using the method’s results, we measured how well the crowd performed in comparison to the expert-based consensus standard.
Experiments and Configuration
We experimented with the method by varying the following dimensions: crowdsourcing platform, compensation amount, and worker quality filters. The first dimension we manipulated was the platform, either CrowdFlower2 and Mechanical Turk3. Next, we compensated workers at either $0.02 per task or $0.06 per task. Finally, we applied quality filters on each platform to filter workers either stringently or not (i.e., low-quality vs. high-quality configuration). Crowdflower provides three levels of worker quality that a requester can specify. Level 1 workers are the lowest quality but provide the fastest response, while Level 3 provides the slowest response but highest quality. On CrowdFlower configurations, we required Level 3 workers for the high-quality configurations. Mechanical Turk awards a Masters certification to workers who have the highest rates of accuracy across a wide variety of tasks. On Mechanical Turk configurations, we required Masters level workers for the high-quality configurations.
Task Factors
In our experiments, we explored the impact of the following four factors:
Task Difficulty The level of challenge in verifying a particular relationship likely affects worker performance. While there is no absolute measure of difficulty, we indirectly captured task difficult by measuring the level of expert agreement on a particular relation. When experts can reach consensus, we consider the task easier than when they cannot reach consensus. Therefore, there are 3 degrees of task difficulty: Pre-Delphi, where experts agreed entirely on a relation in the first round; Post-Delphi, where experts agreed entirely after the Delphi round, and Near-Agreement, where experts agreed with only a super-majority after Delphi. Note that we do not include mere majority agreement because we excluded these relations from the consensus standard.
Definition Quality In previous work, we showed that context (i.e., concept definitions) was critical for a high-performing crowd [32]. To examine this effect in the current study, we asked experts to rate the usefulness of concept definitions during each verification task. We used their response as a proxy for definition quality. Definition quality for a particular relation has three discrete values: none useful, one useful, and two useful.
Worker Ability Simpson and colleague’s aggregation method, which we used to combine worker votes optimally, also estimated the sensitivity and specificity of each worker [33]. We used these estimates to measure average worker ability in each configuration.
Term “Google-ability” Crowd workers often use online search engines to assist with completing a task. Workers may perform better when these search engines provide useful results. To quantify the ease of an online search, we measured the number of search results Google provides for concepts in the verification set4. We performed these searches in February 2015 using an anonymous network connection in an effort to avoid personalized search results.
Analysis
We measured the performance of the crowd-based method by Area Under the Receiver Operating Characteristic curve (AUC). This measure ranges from 0 to 1 and captures how well the methodology performed at identifying the correct and incorrect relationships listed in the expert-based consensus standard at various probability thresholds. Next, to obtain better estimates for AUC, we performed bootstrapping, a process of repeatedly running our experiments by randomly subsampling from the set of relationships. With this bootstrapped distribution of AUCs, we could generalize how the method would perform, on average, with similar datasets. In this study, we performed 10,000 bootstrap iterations for each configuration. We then compared the bootstrapped AUC distributions between various configurations and factors using standard statistical techniques. Specifically, we used t-tests with Benjamini-Hochberg false discovery rate correction when comparing any two configurations. In addition, we used a two-way ANOVA to understand the relative contributions of each factor has on the variability of the AUC. Finally, we used a Wilcoxon rank-sum test, a non-parametric test, for “Google-ability” comparisons because the search count distributions are not normally distributed.
4. Results
We verified a 200 relation subset of the Gene Ontology (GO) in two ways, by experts and by the crowd. Table 1 lists the 16 errors that experts identified by super-majority vote. Note that there is only a small percentage of errors out of 200 relations verified. Further, many of the terms are abstract (e.g., “response to acetate” is generic and difficult to reason about), making the verification task quite challenging. This list of errors serves as the set of incorrect relationships in the consensus standard against which we evaluated the crowd.
Table 1.
Errors in GO subset identified by expert panel
| Child | Parent |
|---|---|
| cellular response to chlorate | cellular response to acid |
| cellular response to fluoride | cellular response to acid |
| cellular response to nitrate | cellular response to acid |
| cellular response to ozone | cellular response to acid |
| extrinsic component of stromal side of plastid inner membrane | extrinsic component of lumenal side of plastid thylakoid membrane |
| positive regulation of mitochondrial membrane permeability involved in apoptotic process | mitochondrial outer membrane permeabilization involved in programmed cell death |
| response to acetate | response to acid |
| response to fluoride | response to acid |
| response to nitrate | response to acid |
| response to nitrite | response to acid |
| response to ozone | response to acid |
| response to chromate | response to transition metal nanoparticle |
| response to manganese ion | response to transition metal nanoparticle |
| response to methylmercury | response to transition metal nanoparticle |
| response to silver ion | response to transition metal nanoparticle |
| response to vanadate(3-) | response to transition metal nanoparticle |
After completing the two survey rounds, experts identified 16 errors in the 200 relationship GO subset we selected. There are three general error categories: acid-related, metal-related, and region-related.
We then used our crowd-backed method to verify the same 200 relations that the experts examined. We ran the methodology with 8 different configurations, faceting on amount paid (high and low), quality filter level (high and low), and platform (Mechanical Turk and CrowdFlower). Table 2 summarizes the performance of the crowd in aggregate (via bootstrapped AUC). We observed a significant difference in performance between all configurations except in two situations. There was no significant difference in AUC between the low-cost, high-quality and high-cost, high-quality configuration on Mechanical Turk. This result indicates that the method’s performance on Mechanical Turk was likely not strongly influenced by rate of reimbursement alone. As an aside, we also calculated the estimated sensitivity and specificity of an average individual worker. Please note, this measure does not describe the crowd’s aggregate performance directly. There was no significant difference in the mean estimated worker sensitivity between the low-cost, low-quality and high-cost, high-quality configuration on CrowdFlower. This indicates that, for this task, the average estimated performance of each individual worker on Crowdflower did not vary (i.e., the average worker was the same in each configuration). While the average worker in each configuration was the same, outliers may account for the variability in the aggregation method’s performance. Therefore, it is possible that higher paying tasks are more likely to attract outliers.
Table 2.
Method performance on verifying GO subset (various configurations)
| Configuration
|
Performance*
|
||||
|---|---|---|---|---|---|
| Platform | Cost | Quality | Mean AUC | Mean Worker Sensitivity | Mean Worker Specificity |
|
|
|
||||
| CrowdFlower | Low | Low | 0.52 | 0.67 | 0.71 |
| High | Low | 0.73 | 0.66 | 0.70 | |
| Low | High | 0.58 | 0.66 | 0.73 | |
| High | High | 0.62 | 0.67 | 0.73 | |
| Mechanical Turk | Low | Low | 0.48 | 0.65 | 0.72 |
| High | Low | 0.60 | 0.65 | 0.73 | |
| Low | High | 0.44 | 0.66 | 0.74 | |
| High | High | 0.44 | 0.62 | 0.72 | |
We measured the performance of the crowd via a bootstrapped AUC and estimated worker sensitivity/specificity in various configurations of platform, cost, and quality. We then compared each configuration to every other configuration to understand whether the performance varied significantly.
Performance on all configuration pairs differed significantly except:
Sensitivity CrowdFlower Low-Cost, Low-Quality vs. CrowdFlower High-Cost, High-Quality
AUC Mechnical Turk Low-Cost, High-Quality vs Mechanical Turk High-Cost, High-Quality
Next, we examined how the crowd performed on subsets of relations. In particular, we stratified relations by degree of expert agreement and by usefulness of the definitions (as rated by the experts) provided for each relation. Table 3 shows the breakdown of the number of relations that fall into the stratified subsets. Note that some strata (e.g., relationships with two useful definitions and near agreement by experts) contain very few relations.
Table 3.
Number of relationships stratified by expert agreement and definition usefulness
| Not Useful | One Useful | Two Useful | All | |
|---|---|---|---|---|
| Pre-Delphi | 81 | 28 | 15 | 124 |
| Post-Delphi | 19 | 10 | 3 | 32 |
| Near-Agreement | 14 | 7 | 4 | 25 |
| All | 114 | 45 | 22 | 181 |
We stratified the selected 200 relationships by two dimensions: (1) expert agreement (based on the Delphi rounds), which served as a proxy for task difficulty where less expert agreement implied higher task difficulty, and (2) expert-rated definition utility, which served as a proxy for definition quality for a relationship where the greater number of useful concept definitions for a relationship implied higher task definition quality. Note that some strata contain very few relationships. (For row label explanation see ‘Task Difficulty’ on page 12. For column label explanation see ‘Definition Quality’ on page 13.)
We then measured the mean bootstrapped AUC of the crowd-backed method on subsets of relationships stratified by expert agreement and definition usefulness. Doing so enabled us to determine where and why crowd performance varied. Table 4 provides the results of this subset analysis. Note that worker performance varies strikingly between various strata. For the majority of configurations, worker performance in each stratum differed significantly from other strata. Further, via ANOVA, we found that definition usefulness and expert agreement along with the interaction of those two dimensions all significantly affect the variability in AUC. This interaction indicates that neither definition usefulness nor the level agreement alone account for the variability of the method’s performance.
Table 4.
Stratified analysis of crowd performance (Mean AUC)
| (a) CrowdFlower: low-cost, low-quality
| ||||
|---|---|---|---|---|
| Not Useful | One Useful | Two Useful | All | |
| Pre-Delphi | 0.08 | 0.13 | ||
| Post-Delphi | 0.53 | 0.47 | ||
| Near-Agreement | 0.78 | 1.00 | 0.63 | |
| All | 0.57 | 0.07 | 0.98 | 0.52 |
| (b) CrowdFlower: high-cost, low-quality
| ||||
|---|---|---|---|---|
| Not Useful | One Useful | Two Useful | All | |
| Pre-Delphi | 0.90 | 0.94 | ||
| Post-Delphi | 0.73 | 0.74 | ||
| Near-Agreement | 0.52 | 1.00 | 0.66 | |
| All | 0.70 | 0.91 | 1.00 | 0.73 |
| (c) CrowdFlower: low-cost, high-quality
| ||||
|---|---|---|---|---|
| Not Useful | One Useful | Two Useful | All | |
| Pre-Delphi | 0.84 | 0.79 | ||
| Post-Delphi | 0.74 | 0.73 | ||
| Near-Agreement | 0.49 | 0.31 | 0.51 | |
| All | 0.56 | 0.84 | 0.25 | 0.58 |
| (d) CrowdFlower: high-cost, high-quality
| ||||
|---|---|---|---|---|
| Not Useful | One Useful | Two Useful | All | |
| Pre-Delphi | 0.38 | 0.31 | ||
| Post-Delphi | 0.53 | 0.53 | ||
| Near-Agreement | 0.55 | 0.07 | 0.70 | |
| All | 0.63 | 0.35 | 0.43 | 0.62 |
| (e) Mechanical Turk: low-cost, low-quality
| ||||
|---|---|---|---|---|
| Not Useful | One Useful | Two Useful | All | |
| Pre-Delphi | 0.65 | 0.62 | ||
| Post-Delphi | 0.41 | 0.40 | ||
| Near-Agreement | 0.38 | 0.11 | 0.47 | |
| All | 0.49 | 0.63 | 0.35 | 0.48 |
| (f) Mechanical Turk: high-cost, low-quality
| ||||
|---|---|---|---|---|
| Not Useful | One Useful | Two Useful | All | |
| Pre-Delphi | 0.60 | 0.68 | ||
| Post-Delphi | 0.50 | 0.55 | ||
| Near-Agreement | 0.41 | 0.43 | 0.62 | |
| All | 0.58 | 0.67 | 0.70 | 0.60 |
| (g) Mechanical Turk: low-cost, high-quality
| ||||
|---|---|---|---|---|
| Not Useful | One Useful | Two Useful | All | |
| Pre-Delphi | 0.32 | 0.42 | ||
| Post-Delphi | 0.36 | 0.39 | ||
| Near-Agreement | 0.57 | 0.20 | 0.50 | |
| All | 0.44 | 0.40 | 0.58 | 0.44 |
| (h) Mechanical Turk: high-cost, high-quality
| ||||
|---|---|---|---|---|
| Not Useful | One Useful | Two Useful | All | |
| Pre-Delphi | 0.30 | 0.33 | ||
| Post-Delphi | 0.48 | 0.48 | ||
| Near-Agreement | 0.37 | 0.06 | 0.38 | |
| All | 0.45 | 0.31 | 0.44 | 0.44 |
All pairs significant except Near-Agreement, Two Useful—All, Two Useful
For each configuration (shown per subtable), we investigated worker performance on the stratified relationships shown in Table 3 (See it for row and column definitions). We measured the crowd performance in those strata by bootstrapped AUC. Next, we performed a Two-Way ANOVA to measure the effect that expert agreement, definition utility, and their interaction have on AUC. In addition, within each configuration, we compared each stratum pairwise to understand where crowd performance differed significantly between those strata.
All pairs significant except: Pre-Delphi, One Useful—All, All
All pairs significant except: All, Two Useful—All, All Task difficulty, definition quality, and their interaction have a significant effect on crowd AUC for every configuration (p<0.05 via Two-Way ANOVA). Crowd AUC between each stratum is significantly different except where noted in the subtable footnote. Blanks indicate there is not at least one correct and incorrect relationship and therefore AUC is incalculable.
While expert agreement is useful for identifying where the crowd will perform well, their agreement is available only in a controlled experiment. A typical application of our method will not have expert comparison. Therefore, we developed an alternative method of quantifying task difficulty by considering the number of results that a Google search will return for a particular term. Figure 6 shows the empirical cumulative distribution of the number of search results for concepts referenced in the GO and SNOMED CT relations verified. Note that there is a marked difference in the number of search results between GO and SNOMED CT in addition to a large difference between search results when the search term is quoted versus unquoted.
Figure 6. Empirical cumulative distribution of Google search results for concepts referenced in verification task.
One possible avenue for the difference in crowd performance is the ease of an online search. As a proxy for ease of search, we ran Google searches for the concepts in the GO and SNOMED CT relationships 4and recorded the number of search results per term. We show these counts as an empirical cumulative distribution.
We then measured whether the search distributions differed significantly from one another. Table 5 describes this comparison. Whether searching with a quoted string or not, there was a significant difference between the number of Google search results. Further, SNOMED CT terms returned a higher number of search results when unquoted. However, when quoted, SNOMED CT terms returned fewer results. This disparity is likely due to the construction of SNOMED CT Fully Specified Names, which do not reflect written or spoken vernacular language. For example, the term “response to acid” from GO likely occurs exactly as written in free text, while the term “Cellulitis and abscess of buttock (disorder)” from SNOMED CT is less likely to occur exactly as written in free text, particularly due to the parenthetical component.
Table 5.
Median number of Google search results for concepts in verification task
| Search Type | Ontology | Median search results* |
|---|---|---|
| Quoted | Gene Ontology | 6125 |
| SNOMED CT | 780 | |
| Unquoted | Gene Ontology | 450000 |
| SNOMED CT | 723500 |
To understand whether the number of search results available for GO concepts and SNOMED CT concepts differed significantly, we ran a Wilcoxon rank-sum test.
There was a statistically significant difference in number of search results between GO and SNOMED CT for both quoted and unquoted searches (p<0.05)
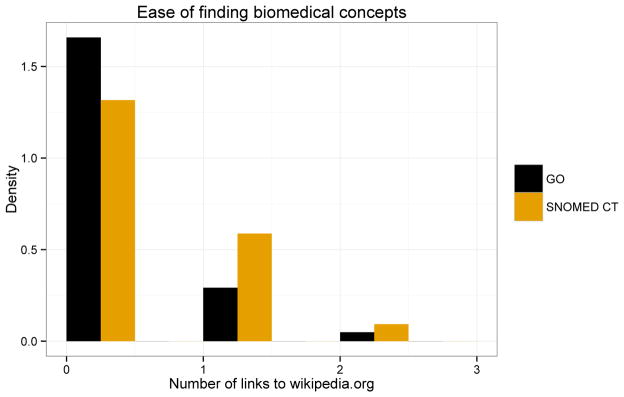
Finally, we honed in on the variability of Wikipedia search results between GO and SNOMED CT. Wikipedia is an information rich source that many crowd workers likely visit while completing tasks. Figure 7 shows the distribution of Wikipedia pages in Google searches results. We observed that SNOMED CT concepts have a greater number of Wikipedia results on average, than do GO concepts – a similar trend to the total number of search results returned for that concept.
Figure 7. Distribution of Wikipedia pages in Google search results for concepts referenced in verification task.
The number of search results does not necessarily indicate useful search results. We captured the difference in useful search results by the number of Wikipedia pages contained within the first page of a Google search result. Note that when searching for concepts in GO or SNOMED CT, there is a considerable difference between the number of Wikipedia pages.
5. Discussion
Experts identified 16 errors in the GO subset, 8% of the relationships they verified. The GO editors have already corrected acid-related errors independently, indicating the errors are indeed “real”. Recall that in our earlier SNOMED CT work, experts identified more than twice as many errors. It is important to note that our sampling of both GO and SNOMED CT likely selected for highly-curated non-trivial entailments. In GO, we selected relationships where concepts had textual definitions, thus requiring a minimum level of curation. In SNOMED CT, we selected relationships that contained concepts that were frequently used in-practice (via CORE subset). Therefore, if we had performed the study on any other, less-curated subset, the error rate would likely be higher. The errors in GO fell into three common categories: unclear definition of acid, unclear definition of metal, and lack of regional clarity for cellular components. For the acid and metal errors, the cause of errors appears to be ambiguity in concept naming conventions. For the regional clarity errors, the cause appears to be subtly incorrect logical definitions (as were many of SNOMED CT errors).
The crowd-based method’s performance in identifying these 16 errors was highly variable, ranging from a mean AUC of 0.73 on CrowdFlower with high compensation to 0.44 on some Mechanical Turk configurations. Because these numbers are bootstrapped, we are confident these results are not random. In certain configurations and certain subsets, the crowd performed perfectly (an AUC of 1). However, they performed rather poorly in others. The highly-paid (i.e., $0.03 per task), low-quality (i.e., no quality filter for worker ability) Mechanical Turk and CrowdFlower configurations show a clear trend – as task difficulty decreases and definition quality increases, workers perform better. This trend is not consistent in all of the data, unfortunately. One potential reason why these configurations exhibited better performance is that the market focuses on highly-paid tasks with low entry requirements. Therefore, platform veterans (i.e., well performing workers) are more likely to complete such tasks when presented in such a configuration. In summary, the crowd does not perform nearly as well on GO as they did on SNOMED CT, but they may still perform well on particular kinds of relationships.
There are several differences between this study and the SNOMED CT study that might contribute to the variability in crowd performance. First, the terms in GO are less frequent in an Internet search (Figure 6). Thus, if workers rely on Google for help with completing the task, they are less likely to find the results they need. If one construes the verification task as a task of understanding and structuring free text knowledge available on the Internet, then workers maybe be less able to succeed because GO has fewer search results. Second, the GO terms themselves are more esoteric than SNOMED CT. For example, workers likely had some familiarity with the anatomy terms in SNOMED CT but have not encountered phrases like “Stromal side of the thylakoid membrane” unless they had advanced training in biology. Finally, the error rate is half that of SNOMED CT. We speculate that this reduced error rate biases workers to hesitate when they see a potential error and that it biases the aggregation method because there is a strong statistical prior against an error occurring. The level of curation in GO is much higher than that of SNOMED CT – in particular, GO is much smaller and has a more active maintenance and Q/A model. The difference in curation may account for the reduced error rate. In short, it appears that verifying GO is simply more difficult than verifying SNOMED CT. The results suggest that portions of the Gene Ontology sit at the boundary of the crowd’s capabilities to complete expert-level, knowledge-intensive tasks.
The difficulty of verifying GO and the crowd’s poor performance underscores the importance of intelligent worker and task selection. As we have shown in previous work, and as some configurations in this experiment showed, the crowd performs best with appropriate context (i.e., definitions) and with easier tasks [32]. Therefore, it is important to be able to quantify, a priori, how difficult a task is without knowing the correct answer. Considering the change in search results between GO and SNOMED CT, we propose using the number of search results as a predictor of task difficulty. In addition, it is important to have high-quality definitions. When a relationship’s concepts lack definitions, one potential solution is to use crowdsourcing to assist with generating them. Such a method must exercise caution, as a crowd worker could generate a poor definition and later verify a relationship as “correct” based on that poor definition. Heuristics and metrics to assess definition quality would be essential to a combined define and verify system. In a real-world, crowd-based ontology verification situation, an ontology developer could first measure programatically the number of Internet search results for a given relationship. Only when the number of search results (or Wikipedia entries) exceeds some threshold would the ontology developer send out the task to the crowd. She would route the remaining relationships to an expert or verify them herself. In such a setup, the crowd is an ontology engineering assistant, rather than an independently operating substitute expert. Here, the crowd would complete easy tasks, reducing the load of an ontology engineer and would return tasks for review for which they are not confident (i.e., the probability of an error is ~0.5).
When verifying GO, the crowd performed poorly. Indeed, it appears the crowd alone cannot address all the challenges of improving ontology engineering methods. The crowd likely is best included in part of a larger system, much like the assistant described above. We call this system a “group-sourced” ontology development environment. Here, the crowd, the computer, and expert work in concert. Each contributes their strengths, completing tasks best suited to them. For example, suppose an ontology engineer is developing a large, logically-complex biomedical ontology. In the background, a computer-based agent would search for areas in the ontology with potential errors using a battery of quality heuristics. Once the agent identifies candidate regions in the ontology, it would determine, by applying another set of rules (e.g., the number of search results as a threshold), which relationships to bring to the attention of the human developer and which it could verify via crowdsourcing. Later, the agent would gather, aggregate, and present the crowdsourced results to the expert. The agent would learn continually about the engineer based on her responses to its tasks and tailor its actions to best fit her requirements and expectations. Further, the agent would retain a customized crowd workforce who perform best at the verification task. This workflow is applicable in more ontology engineering than just verification. It could also help with ontology generation, mapping, and alignment. We envision such a system integrated into our collaborative Protégé ontology development environment [37]. Here, multiple experts collaborate together in an online tool to construct an ontology, typically focusing on a component in which they specialize. The “group-sourcing” system outlined above would enhance the collaborative Protégé tool greatly, providing its users with additional ways to engineer and evaluate the ontologies they build by leveraging the computer-based agent and the crowd.
To move toward such a system, the next step is to perform a large-scale application of the crowd-based verification method. Our studies thus far have been limited by expert availability and resource availability. A larger analysis would focus on a single, complete ontology development cycle (from editing to release) in collaboration with an organization that produces a a biomedical ontology (e.g., GO Consortium). This study would serve to confirm the capability of the methodology and to pilot the basics of an integrated “group-sourcing” system. There are two potential approaches we propose: (1) a split trial, in which different engineers complete the same cycle with or without the assistance of the crowdsourcing method, or (2) a retrospective study, in which engineers complete the cycle with the crowd-backed method and they compare their results to those of prior releases. In either, we would measure how the ontologies change, determine error rates via third party review, record the resource requirements, and survey the user experience. Our results thus far have been encouraging and we look forward to seeing how the crowd scales.
6. Conclusions
As ontology use increases in biomedicine, its important to minimize errors. Current methods that detect errors have two main limitations: scalability and accuracy. In prior work, we developed a crowdsourcing-based method that overcomes these limitations and show it performed well when verifying SNOMED CT. Here, we applied that same method, in various configurations (cost, quality, and platform), to verify the Gene Ontology. On the whole, the crowd did not perform as well as they did on SNOMED CT. However, in certain configurations, the crowd performed reasonably well, particularly on tasks where experts rated the definitions as useful and reached early agreement. Further investigation into where the boundary between crowd and expert lies is certainly warranted. These results suggest that the crowd is not a panacea, but instead a powerful tool that performs best when working with the appropriately selected tasks (i.e., the ones with good context that are not overly difficult). Considering that, we outlined a system in which a computational agent, the crowd, and experts all work together to construct high-quality, error-free ontologies.
Highlights.
Experts identified 16 errors, mainly related to acids and metals
Definition quality and task difficulty affect crowd performance significantly
The number of Internet search results could help identify “easy” crowd tasks
The crowd fits best as an assistant, helping experts with their verification tasks
Acknowledgments
This work was supported by the National Institute of General Medical Sciences grant number GM086587, by the National Center for Biomedical Ontology, supported by the National Human Genome Research Institute, the National Heart, Lung, and Blood Institute, and the National Institutes of Health Common Fund grant number HG004028, and by the National Library of Medicine Informatics Training grant number LM007033.
Footnotes
Publisher's Disclaimer: This is a PDF file of an unedited manuscript that has been accepted for publication. As a service to our customers we are providing this early version of the manuscript. The manuscript will undergo copyediting, typesetting, and review of the resulting proof before it is published in its final citable form. Please note that during the production process errors may be discovered which could affect the content, and all legal disclaimers that apply to the journal pertain.
References
- 1.Staab S, Studer R. Handbook on Ontologies. 2. Springer-Verlag New York Inc; 2009. URL http://books.google.com/books?hl=en&lr=&id=W6ZNcAolVbwC&oi=fnd&pg=PA1&dq=Handbook+on+Ontologies&ots=fAHz8ROP-D&sig=QfPb_USrS1t-No4H8jPPCmVoX-w. [Google Scholar]
- 2.Singhal A. Official Google Blog. May, Introducing the knowledge graph: things, not strings. [Google Scholar]
- 3.Bodenreider O, Stevens R. Bio-ontologies: current trends and future directions. Briefings in bioinformatics. 2006;7(3):256–274. doi: 10.1093/bib/bbl027. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Bodenreider O. Biomedical ontologies in action: role in knowledge management, data integration and decision support. Yearbook of medical informatics. 2008:67–79. me08010067[pii] [PMC free article] [PubMed] [Google Scholar]
- 5.Rubin DL, Shah NH, Noy NF. Biomedical ontologies: a functional perspective. Briefings in Bioinformatics. 2008;9(1):75–90. doi: 10.1093/bib/bbm059. [DOI] [PubMed] [Google Scholar]
- 6.Hunter L, Lu Z, Firby J, Baumgartner WA, Johnson HL, Ogren PV, Cohen KB. OpenDMAP: an open source, ontology-driven concept analysis engine, with applications to capturing knowledge regarding protein transport, protein interactions and cell-type-specific gene expression. BMC bioinformatics. 2008;9(1):78. doi: 10.1186/1471-2105-9-78. URL http://www.biomedcentral.com/1471-2105/9/78. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Hoehndorf R, Dumontier M, Gennari JH, Wimalaratne S, de Bono B, Cook DL, Gkoutos GV. Integrating systems biology models and biomedical ontologies. BMC systems biology. 2011;5(1):124. doi: 10.1186/1752-0509-5-124. URL http://www.biomedcentral.com/1752-0509/5/124. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Ashburner M, Ball CA, Blake JA, Botstein D, Butler H, Cherry JM, Davis AP, Dolinski K, Dwight SS, Eppig JT, Harris MA, Hill DP, Issel-Tarver L, Kasarskis A, Lewis S, Matese JC, Richardson JE, Ringwald M, Rubin GM, Sherlock G. Gene Ontology: tool for the unification of biology. Nature genetics. 2000;25(1):25–29. doi: 10.1038/75556. URL http://www.ncbi.nlm.nih.gov/pmc/articles/PMC3037419/http://www.ncbi.nlm.nih.gov/pmc/articles/PMC3037419/pdf/nihms-269796.pdf. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Khatri P, Sirota M, Butte AJ. Ten years of pathway analysis: current approaches and outstanding challenges. PLoS computational biology. 2012;8(2):e1002375. doi: 10.1371/journal.pcbi.1002375. URL http://www.pubmedcentral.nih.gov/articlerender.fcgi?artid=3285573&tool=pmcentrez&rendertype=abstract. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Musen MA, Noy NF, Shah NH, Whetzel PL, Chute CG, Storey M-A, Smith B. The National Center for Biomedical Ontology. Journal of American Medical Informatics Association. 2012;19:190–195. doi: 10.1136/amiajnl-2011-000523. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Rector AL, Brandt S, Schneider T. Getting the foot out of the pelvis: modeling problems affecting use of SNOMED CT hierarchies in practical applications. Journal of the American Medical Informatics Association. 2011;18(4):432–440. doi: 10.1136/amiajnl-2010-000045. URL http://jamia.bmj.com/cgi/doi/10.1136/amiajnl-2010-000045. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Ceusters W, Smith B, Goldberg L. A terminological and ontological analysis of the NCI Thesaurus. Methods of information in medicine. 2005;44(4):498. URL http://www.ncbi.nlm.nih.gov/pubmed/16342916http://citeseerx.ist.psu.edu/viewdoc/summary?doi=10.1.1.90.4270http://ontology.buffalo.edu/medo/NCIT.pdfciteulike-article-id:6080807http://view.ncbi.nlm.nih.gov/pubmed/16342916. [PubMed] [Google Scholar]
- 13.Ceusters W. Applying Evolutionary Terminology Auditing to the Gene Ontology. Journal of biomedical informatics. 2009:1–41. doi: 10.1016/j.jbi.2008.12.008. URL http://www.sciencedirect.com/science/article/pii/S1532046408001524. [DOI] [PubMed]
- 14.Mortensen JM, Minty EP, Januszyk M, Sweeney TE, Rector AL, Noy NF, Musen MA. Using the wisdom of the crowds to find critical errors in biomedical ontologies: a study of SNOMED CT. Journal of the American Medical Informatics Association. doi: 10.1136/amiajnl-2014-002901. URL http://jamia.bmj.com/content/early/2014/10/23/amiajnl-2014-002901.abstract. [DOI] [PMC free article] [PubMed]
- 15.Geller J, Perl Y, Halper M, Cornet R. Special issue on auditing of terminologies. Journal of biomedical informatics. 2009;42(3):407–11. doi: 10.1016/j.jbi.2009.04.006. URL http://www.sciencedirect.com/science/article/pii/S1532046409000598http://www.ncbi.nlm.nih.gov/pubmed/19465342. [DOI] [PubMed] [Google Scholar]
- 16.Gangemi A, Catenacci C, Ciaramita M, Lehmann J. The Semantic Web: Research and Applications, Vol. 4011 LNCS. Springer; 2006. Modelling ontology evaluation and validation; pp. 140–154. [DOI] [Google Scholar]
- 17.Lozano-Tello A, Gómez-Pérez A. Ontometric: A method to choose the appropriate ontology. Journal of Database Management. 2004;2(15):1–18. [Google Scholar]
- 18.Tartir S, Arpinar IB, Moore M, Sheth AP, Aleman-meza B. OntoQA: Metric-based ontology quality analysis. IEEE Workshop on Knowledge Acquisition from Distributed, Autonomous, Semantically Heterogeneous Data and Knowledge Sources. 2005;9 [Google Scholar]
- 19.Gruber TR. Tech Rep. Vol. 56. Knowledge Systems Laboratory, Stanford University; 1993. Toward Principles for the Design of Ontologies Used for Knowledge Sharing. URL http://www.sciencedirect.com/science/article/pii/S1071581985710816. [DOI] [Google Scholar]
- 20.Dietrich J, Elgar C. A Formal Description of Design Patterns Using OWL. 2005 Australian Software Engineering Conference. 2005:243–250. doi: 10.1109/ASWEC.2005.6. URL http://ieeexplore.ieee.org/lpdocs/epic03/wrapper.htm?arnumber=1402019. [DOI]
- 21.Aranguren MEn, Antezana E, Kuiper M, Stevens R. Ontology Design Patterns for bio-ontologies: a case study on the Cell Cycle Ontology. BMC bioinformatics. 2008;9(Suppl 5):S1. doi: 10.1186/1471-2105-9-S5-S1. URL http://www.pubmedcentral.nih.gov/articlerender.fcgi?artid=2367624&tool=pmcentrez&rendertype=abstract. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Gangemi A. Ontology Design Patterns for Semantic Web Content. 4th International Semantic Web Conference (ISWC2005); Galway, Ireland: Springer; 2005. pp. 262–276. URL http://www.springerlink.com/index/F513071T4477H2W2.pdf. [Google Scholar]
- 23.Quinn AJ, Bederson BB. Human computation: A Survey and Taxonomy of a Growing Field. Proceedings of the 2011 annual conference on Human factors in computing systems - CHI ’11; Vancouver, BC: ACM; 2011. pp. 1403–1412. URL http://portal.acm.org/citation.cfm?doid=1978942.1979148$\delimiter”026E30F$nhttp://www.alexquinn.org/papers/HumanComputation,ASurveyandTaxonomyofaGrowingField(CHI2011).pdf. [DOI] [Google Scholar]
- 24.Lintott CJ, Schawinski K, Slosar A, Land K, Bamford S, Thomas D, Raddick MJ, Nichol RC, Szalay A, Andreescu D, Murray P, Vandenberg J. Galaxy Zoo: morphologies derived from visual inspection of galaxies from the Sloan Digital Sky Survey. Monthly Notices of the Royal Astronomical Society. 2008;389(3):1179–1189. doi: 10.1111/j.1365-2966.2008.13689.x. URL http://dx.doi.org/10.1111/j.1365-2966.2008.13689.x. [DOI] [Google Scholar]
- 25.Cooper S, Khatib F, Treuille A, Barbero J, Lee J, Beenen M, Leaver-Fay A, Baker D, Popovi Z. Predicting protein structures with a multiplayer online game. Nature. 2010;466(7307):756–760. doi: 10.1038/nature09304. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Demartini G, Difallah DE, Cudré-Mauroux P. ZenCrowd: leveraging probabilistic reasoning and crowdsourcing techniques for large-scale entity linking. 21st World Wide Web Conference WWW; 2012; Lyon, France. 2012. pp. 469–478. [Google Scholar]
- 27.MacLean DL, Heer J. Identifying medical terms in patient-authored text: a crowdsourcing-based approach. Journal of the American Medical Informatics Association. doi: 10.1136/amiajnl-2012-001110. URL http://jamia.bmj.com/content/early/2013/05/04/amiajnl-2012-001110.full. [DOI] [PMC free article] [PubMed]
- 28.Siorpaes K, Hepp M. Games with a Purpose for the Semantic Web. IEEE Intelligent Systems. 2008;23(3):50–60. doi: 10.1109/MIS.2008.45. URL http://ieeexplore.ieee.org/xpl/articleDetails.jsp?arnumber=4525142. [DOI] [Google Scholar]
- 29.Siorpaes K, Hepp M. OntoGame: Weaving the Semantic Web by Online Games. In: Bechhofer S, Hauswirth M, Hoffmann J, Koubarakis M, editors. The Semantic Web: Research and Applications SE - 54, Vol. 5021 of Lecture Notes in Computer Science. Springer; Berlin Heidelberg: 2008. pp. 751–766. URL http://dx.doi.org/10.1007/978-3-540-68234-9_54. [DOI] [Google Scholar]
- 30.Sarasua C, Simperl E, Noy NF. CrowdMAP: Crowdsourcing Ontology Alignment with Microtasks. 11th International Semantic Web Conference (ISWC); Boston, MA: Springer; 2012. [Google Scholar]
- 31.Noy NF, Mortensen JM, Alexander PR, Musen MA. Mechanical Turk as an Ontology Engineer? Using Microtasks as a Component of an Ontology Engineering Workflow. Web Science. 2013 [Google Scholar]
- 32.Mortensen JM, Alexander PR, Noy NF, Musen MA. Crowd-sourcing Ontology Verification. International Conference on Biomedical Ontologies. 2013 [Google Scholar]
- 33.Simpson E, Roberts S, Psorakis I, Smith A. Dynamic Bayesian Combination of Multiple Imperfect Classifiers. 2012 arXiv. 35arXiv:1206.1831. URL http://arxiv.org/abs/1206.1831.
- 34.Mortensen JM, Musen MA, Noy NF. An empirically derived taxonomy of errors in SNOMED CT. AMIA Annual Symposium Proceedings 2014. 2014:899–906. URL http://www.ncbi.nlm.nih.gov/pmc/articles/PMC4419962/ [PMC free article] [PubMed]
- 35.Horridge M, Bechhofer S. The OWL API: A Java API for Working with OWL 2 Ontologies. OWLED. 2009;529:11–21. URL http://www.webont.org/owled/2009/papers/owled2009_submission_29.pdf. [Google Scholar]
- 36.Linstone HA, Turoff M. The Delphi method: techniques and applications. Addison-Wesley; Reading: 1975. [Google Scholar]
- 37.Tudorache T, Noy NF, Tu S, Musen MA. Supporting Collaborative Ontology Development in Protege. 7th International Semantic Web Conference (ISWC 2008); Karlsruhe, Germany. 2008. [Google Scholar]