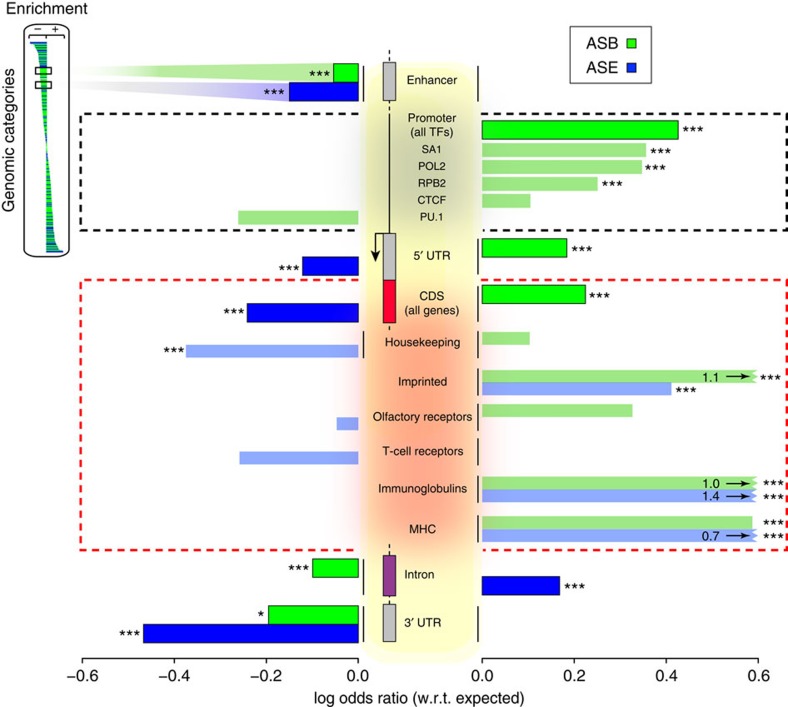
Figure 5. Population-aware 'expanded' enrichment analysis shows that some genomic regions are more inclined to allele-specific regulation.
The ‘expanded' analysis is performed in a population-aware manner, where each control or allele-specific SNV is counted once for each occurrence in an individual. We map variants associated with ASB (green) and ASE (blue) to various categories of genomic annotations, such as CDSs, UTRs, enhancer and promoter regions, to survey the human genome for regions more enriched in allelic behaviour. Using the control non-allele-specific SNVs as the expectation, we compute the log odds ratio for ASB and ASE SNVs separately, via Fisher's exact tests. Bonferroni-corrected: *P<0.05; **P<0.01; ***P<0.001. For each TF in AlleleDB, we also calculate the log odds ratio of ASB SNVs in promoters, providing a proxy of allele-specific regulatory role for each available TF. Genes known to be mono-allelically expressed such as imprinted and MHC genes (CDS regions) are highly enriched for both ASB and ASE SNVs. The actual log odds ratio of ASB SNVs in imprinted genes, both ASB and ASE SNVs in immunoglobulin genes and ASE SNVs for MHC is indicated on the bar.