Figure 1. An unbiased LIN28B 3′UTR-miRNA library screen identifies 28 miRNAs targeting LIN28B.
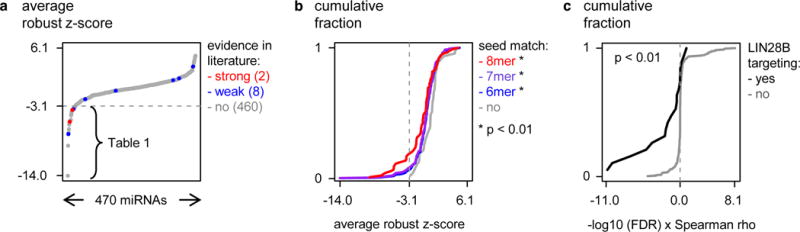
(a) Average robust z-scores are plotted (Y-axis) for 470 tested miRNAs (X-axis). The 28 miRNAs with a z-core of < −3.05 are listed in Table 1. The 10 interactions that have been reported in literature according to miRtarBase, are indicated in red (strong evidence) and blue (weak evidence). (b) Cumulative distribution (Y-axis) of the robust z-scores (X-axis) of miRNAs that respectively have 6-, 7- or 8-mer seed-matches in the LIN28B 3′UTR. P-value represents Kolmogorov-Smirnov test for the distribution of the average robust z-score for miRNAs that have a 6-, 7- or 8-mer seed-match versus miRNAs without seed-match. (c) The cumulative distribution (Y-axis) of the π-value for the correlation between miRNA and LIN28B expression (X-axis) of miRNAs that were identified as either LIN28B-targeting (black) or not LIN28B-targeting (gray). Kolmogorov-Smirnov test, p = 1.4 × 10−3.
