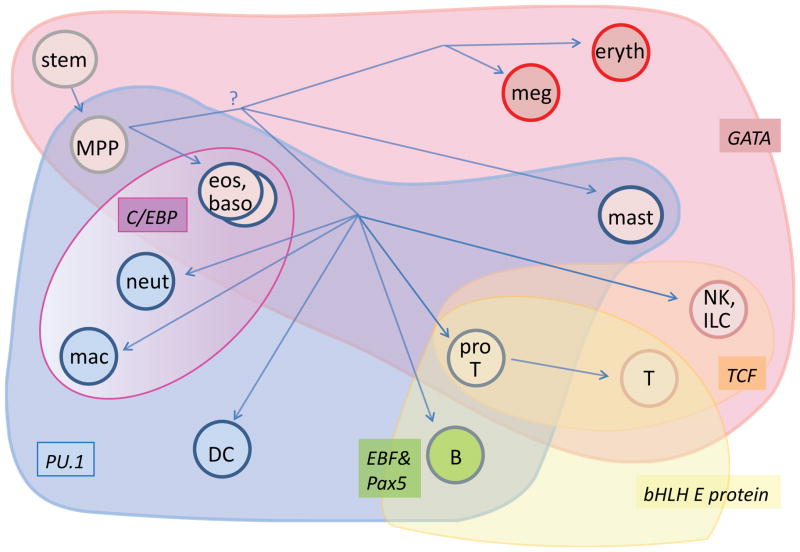
Figure 2.
Broadly combinatorial use of key transcription factors in specifying distinct hematopoietic cell fates. A schematic view is presented of mouse hematopoiesis, superimposed by the overlapping domains of activity of PU.1 (blue), GATA family factors (GATA-1, GATA-2, or GATA-3, red), C/EBP family factors (violet, graded to indicate dosage relative to PU.1), E proteins (E2A and/or HEB, yellow), TCF family factors (TCF-1/LEF-1, orange), and EBF1+Pax5 (B lineage specific, green). Whereas C/EBP family factors do not appear to overlap with the domains of E proteins and TCF-1/LEF-1, and the EBF1+Pax5 combination is restricted to a single cell type, there is extensive variation in the combinations of GATA, PU.1, C/EBP, E proteins, and TCF-1/LEF-1 that are allowed to distinguish the other cell fates shown. Eryth=erythrocyte. Meg=megakaryocyte. Eos=eosinophil. Baso=basophil. Neut=neutrophil. Mac=macrophage. DC=dendritic cell. ILC=innate lymphoid cell. NK=natural killer. ProT= T-cell precursor up to commitment (during period of PU.1 expression).