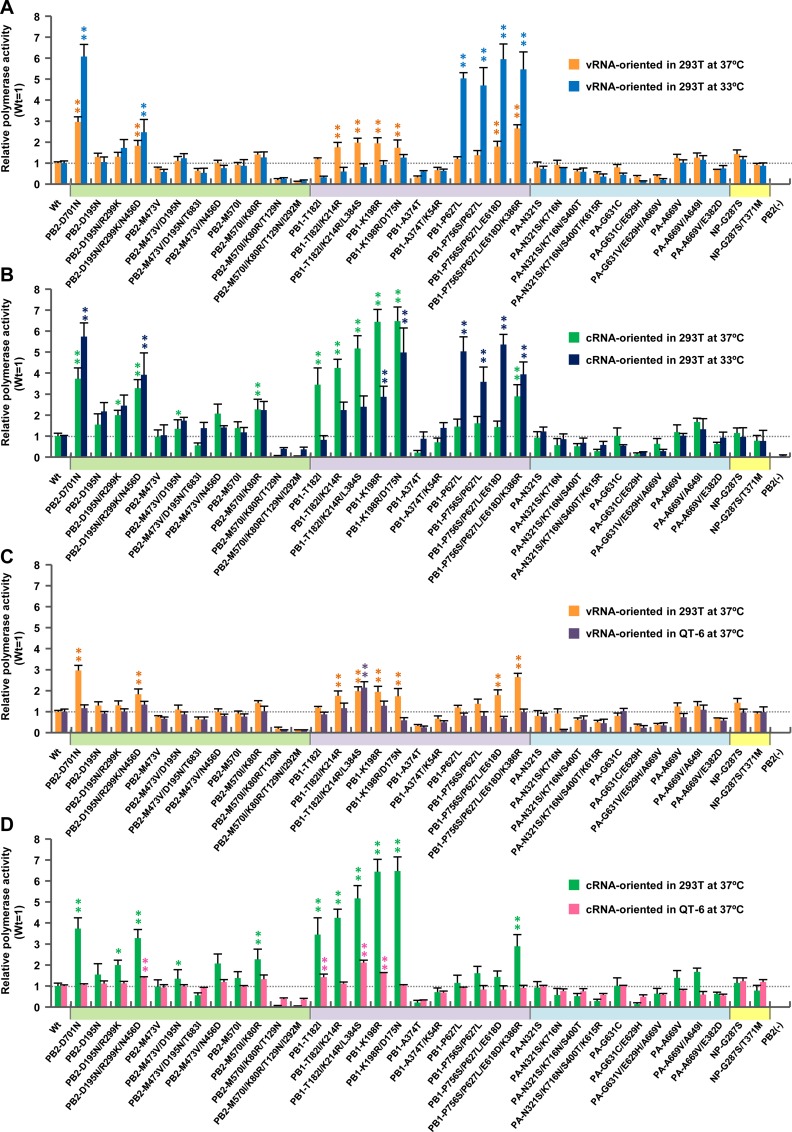
Fig 3. Effect of multiple mutations on polymerase activity.
293T and QT-6 cells were transfected with plasmids expressing EG/D1 PB2, PB1, PA and NP with the indicated multiple mutations, a human or chicken polymerase I-driven plasmid expressing a vRNA-oriented or cRNA-oriented luciferase reporter gene, and a Renilla luciferase-expressing plasmid as an internal control. Luciferase activities were assayed 48 h post-transfection and normalized to the internal Renilla luciferase activity. The data were expressed relative to the results for EG/D1 (wt). (A) Comparison of luciferase activity at 37 and 33°C in vRNA-oriented minigenome assays in 293T cells. (B) Comparison of polymerase activity at 37 and 33°C in cRNA-oriented minigenome assays in 293T cells. (C) Comparison of polymerase activity in 293T and QT6 cells in vRNA-oriented minigenome assays at 37°C. (D) Comparison of polymerase activity in 293T and QT6 cells in cRNA-oriented minigenome assays at 37°C. Each data point is the mean ± SD of three independent experiments. Colors on each x-axis highlight different virus gene segments. Single and double asterisks indicate a P value <0.05 and <0.01, respectively (ANOVA with Tukey’s multiple comparison test). Asterisks for mutations with negative effects on polymerase activity were omitted for clarity.