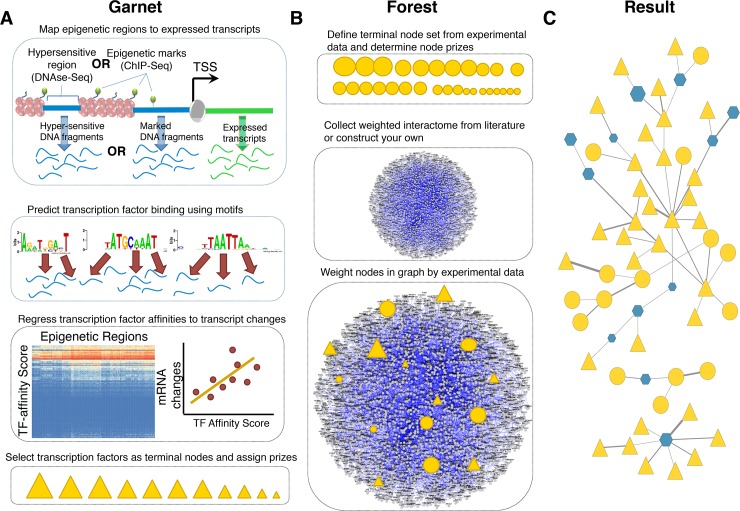
Fig 2.
Summary of Omics Integrator (A) Garnet identifies transcription factors (triangles) associated with mRNA expression changes by incorporating epigenetic changes nearby expressed genes, scanning those regions for putative transcription factor binding sites and then regressing transcription factor affinity scores against gene expression changes. The result is a set of transcription factor candidates and the relative confidence that they are responsible for the observed expression changes. (B) Forest identifies a condition-specific functional sub-network from user data and a confidence-weighted interactome. The network can be composed of protein-protein, protein-metabolite or other interactions. The set of omic hits are composed of the TFs obtained from Garnet (triangles) merged with other types of hits such as differentially expressed proteins, significantly phosphorylated proteins, metabolites, etc. (circles). (C) Finally, the confidence-weighted interactome is integrated with the ‘omic’ hits using the prize-collecting Steiner forest algorithm, where the data is either connected directly or via intermediate nodes, called ‘Steiner nodes’.