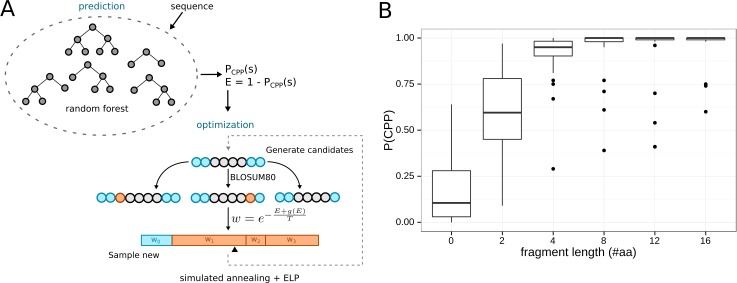
Fig 1. Identification and design of CPP-like sequences.
(A) Methodology used to identify and design CPP-like sequences. First, a random forest predictor is trained on a set of known CPP and non-CPP sequences. The resulting probabilistic model is then used to identify CPP-like sequences and converted into an energy measure (E = 1 –PCPP(S)) for novel peptide design by simulated-annealing optimization (see Materials and Methods). (B) Algorithm performance during the design of CPP-like sequences. 32 random peptide sequences of length 16 were optimized with varying fragment lengths for 1000 iterations. Short fragments of more than 4 amino acids proved sufficient to transform non-CPP sequences to CPP-like sequences.