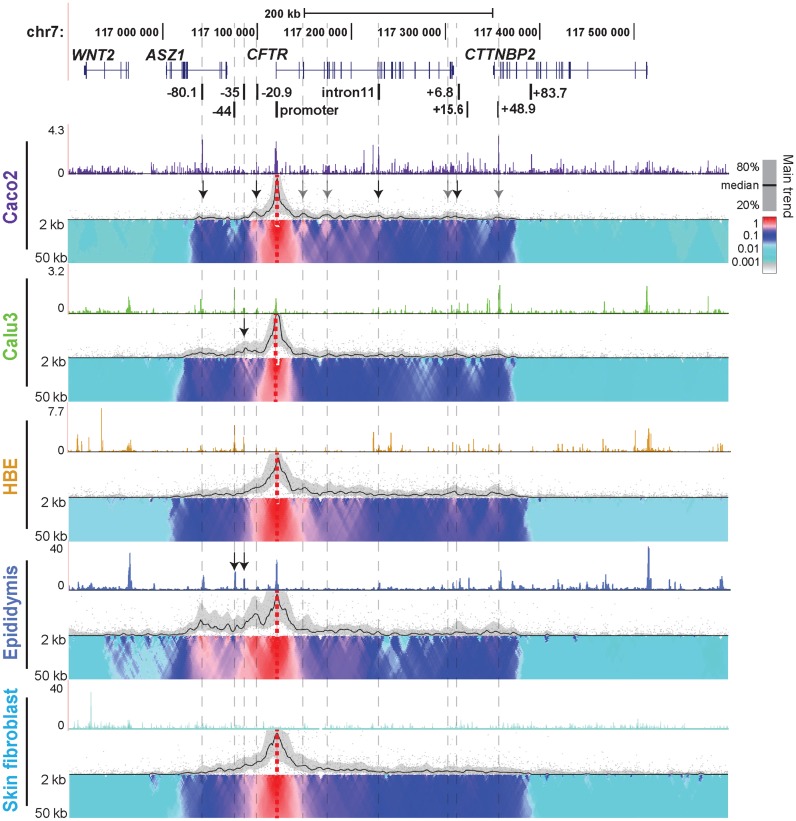
Figure 1.
Cell-type-specific chromatin structure at the CFTR locus: 4C-seq profiles with a CFTR promoter viewpoint. A schematic at the top shows the genomic location of CFTR and adjacent genes on chromosome 7 and below are known cis-regulatory elements for the CFTR locus. The nomenclature represents distance in kb (−) 5′ to the translational start site and (+) 3′ to the last coding base. Open chromatin mapped by DNase-seq and 4C-seq data are shown for Caco2, Calu3, HBE, epididymis and skin fibroblast cells. Representative data from one of the replicates are shown here. DNase-seq data are presented as histograms from screen shots of data sets uploaded to the UCSC genome browser. 4C-seq data are presented in alignment with the DNase-seq data and have two subpanels. The upper panel indicates the main trend of contact profile using a 5-kb window size. Relative interactions are normalized to the strongest point (which is set to 1) within each panel. The lower panel is a domainogram (39), which uses color coded intensity values to show relative interactions with window sizes varying from 2 to 50 kb. Here, red denotes the strongest interactions and dark blue, through turquoise, to gray represent gradually decreasing frequencies. Arrows denote important data features described in the results.