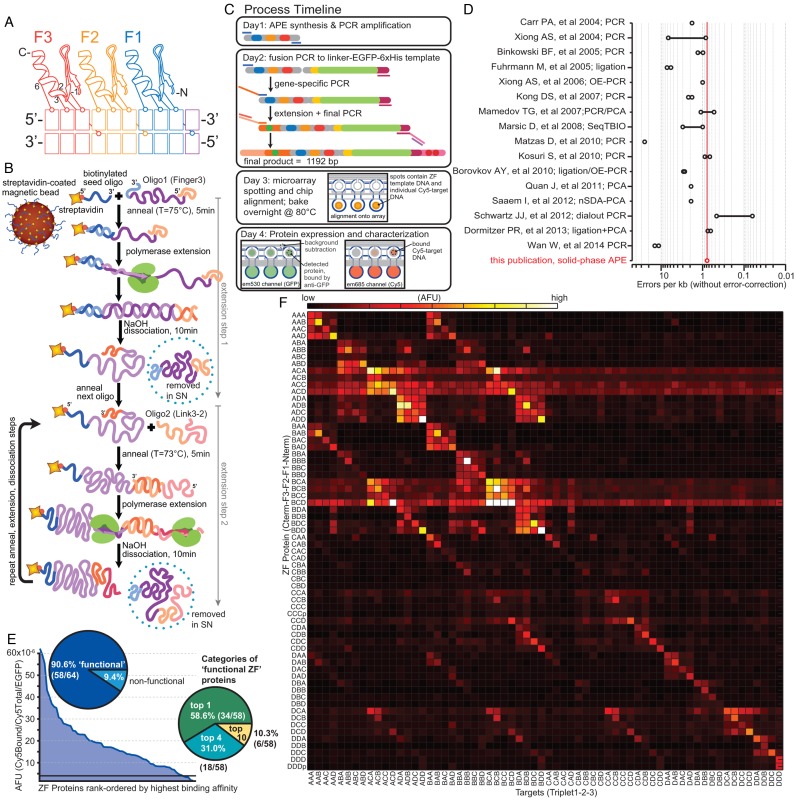
Figure 1.
APE-MITOMI applied to ZF TF module combinatorics. (A) Cartoon model of canonical Cys2His2 ZF TF binding to DNA with residues −1, 2, 3 and 6 of the recognition helix primarily encoding DNA specificity. Residue 2 makes a cross-strand contact, which creates ‘context dependent’ effects. (B) Schematic of the APE solid-phase gene assembly technique, showing assembly through the first two extension steps. (C) Process timeline from gene assembly to protein characterization. (D) Comparison of APE error rate with values from previously published gene assembly techniques. A line between two points indicates a range of error rates from different experimental conditions. (E) Overview of experimental results obtained from combinatoric assembly of ZF TFs demonstrating protein expression and functional DNA binding success rates. (F) Heatmap of relative binding affinities for each assembled ZF TF (y-axis) to 64 predicted consensus DNA targets (x-axis). Protein naming convention indicates ZF domain from C-to-N (F3 to F1), where AAA (Af3Af2Af1) = Zif268, BBB = 37–12, CCC = 92–1, DDD = 158–2 (14); for example, protein ABC = F3 from Zif268, F2 from 37–12, F1 from 92–1; target ABC = Zif268 F3 binding consensus triplet GCG, 37–12 F2 binding consensus GAC, 92–1 F1 binding consensus triplet GCC (5′-GCG GAC GCC). The values represent averages of multiple measurements and the precise number of technical repeats and a histogram thereof are shown in Supplementary Figures S7 and S8, respectively. Oligomer assembly and target sequences are given in Supplementary Tables S7 and S8.