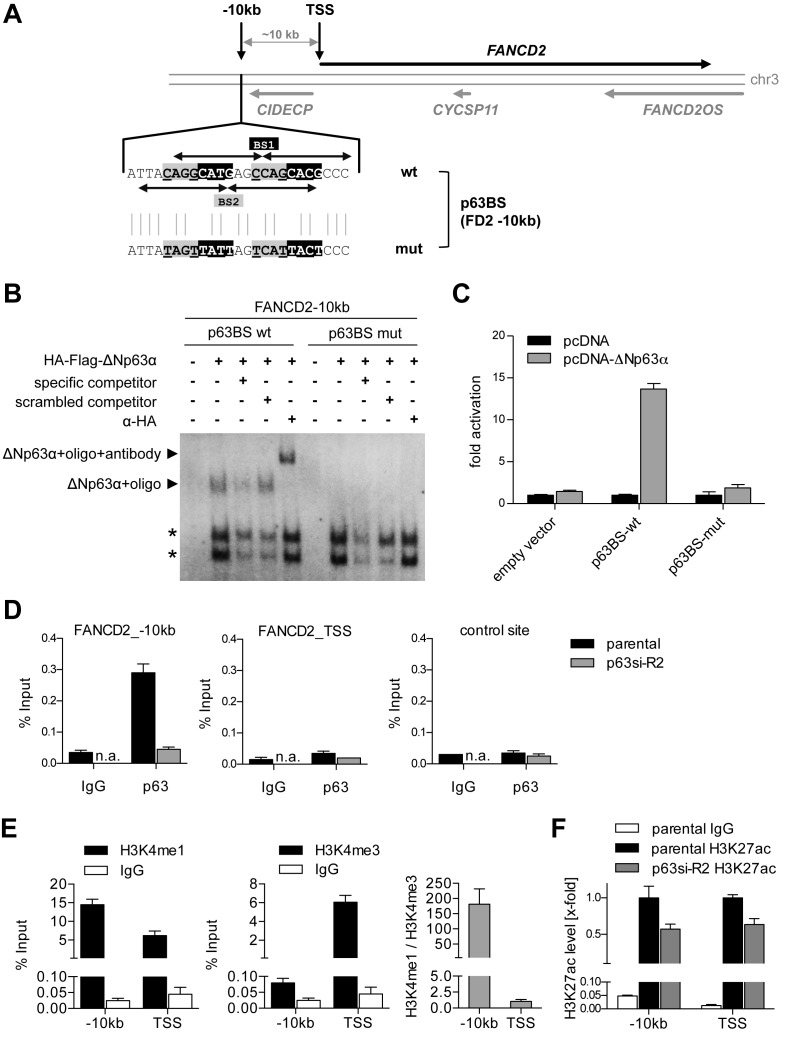
Figure 5.
ΔNp63 binds and regulates an enhancer upstream of FANCD2. (A) Genomic locus of FANCD2 with identified p63 binding site (BS) 10 kb upstream of the FANCD2 transcription start site (TSS). Arrows indicate decameric half-sites of two intertwined p63BS 1 and 2. Mutation of p63BS (p63BS mut) of the central CnnG motif is depicted. (B) EMSA for binding of in vitro translated HA-tagged ΔNp63 to wild-type and mutated FANCD2 −10 kb p63BS. Specificity of the complex was confirmed by competition with unlabeled specific or scrambled competitor and supershift with HA-antibody. Asterisks indicate unspecific complexes. (C) Luciferase reporter assay analyzing ΔNp63-mediated activation of a firefly luciferase via wild-type but not mutated FANCD2 −10 kb p63BS. Bars show relative activation of the luciferase reporter compared to empty vector control (pcDNA) (mean +SD (n = 3)). (D) Chromatin immunoprecipitation (ChIP) demonstrating in vivo binding of endogenous ΔNp63 at the FANCD2 −10 kb but not TSS or negative control region in UT-SCC-74A before (parental) and after depletion of p63 (p63si-R2). Binding is shown as mean % input +SD (n = 2). IgG, negative control ChIP; n.a., not analyzed. (E) Analysis of chromatin state at FANCD2 −10 kb and TSS site by H3K4me1 and H3K4me3 ChIP compared to IgG control. Histone modification is shown as mean % input +SD (n = 2) and H3K4me1/me3 ratio. (F) ChIP analyzing histone H3 lysine 27 acetylation (H3K27ac) as a marker for activity. FANCD2 −10 kb and TSS were evaluated in untreated (parental) and p63-depleted (p63si-R2) UT-SCC-74A. Bars show relative H3K27ac level of % input at −10 kb and TSS region normalized to parental cells (n = 3).