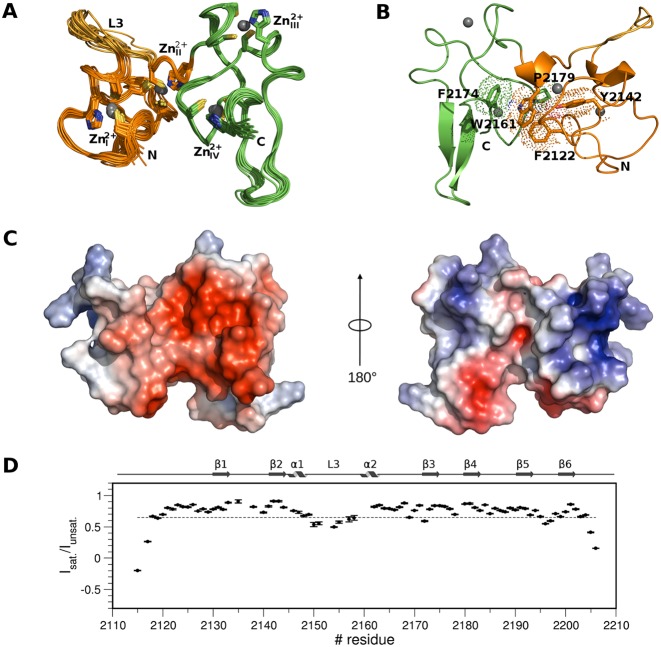
Figure 1.
Solution structure of PHDVC5HCHNSD1. (A) Superposition of the best 15 NMR structures PHDV; the L3 loop and C5HCH are coloured in orange, gold and green, respectively. Zn2+ binding residues and Zn2+ ions are represented in sticks and spheres, respectively. (B) Cartoon representation of PHDVC5HCHNSD1 highlighting elements of secondary structure. Residues forming the domain hydrophobic core at the domains interface are shown in sticks and dotted space-filling representation. Zn2+ ions are represented in spheres. Amino acids in this and in the following Figures are numbered according to the human NSD1 sequence. (C) Electrostatic surface potential of PHDVC5HCHNSD1. The structures on the left and on the right are represented in the same orientation as in (A) and (B), respectively. (D) Backbone dynamics of PHDVC5HCHNSD1. Dotted line indicates the {1H}–15N heteronuclear NOE value threshold of 0.65. Elements of secondary structure are indicated on the top.