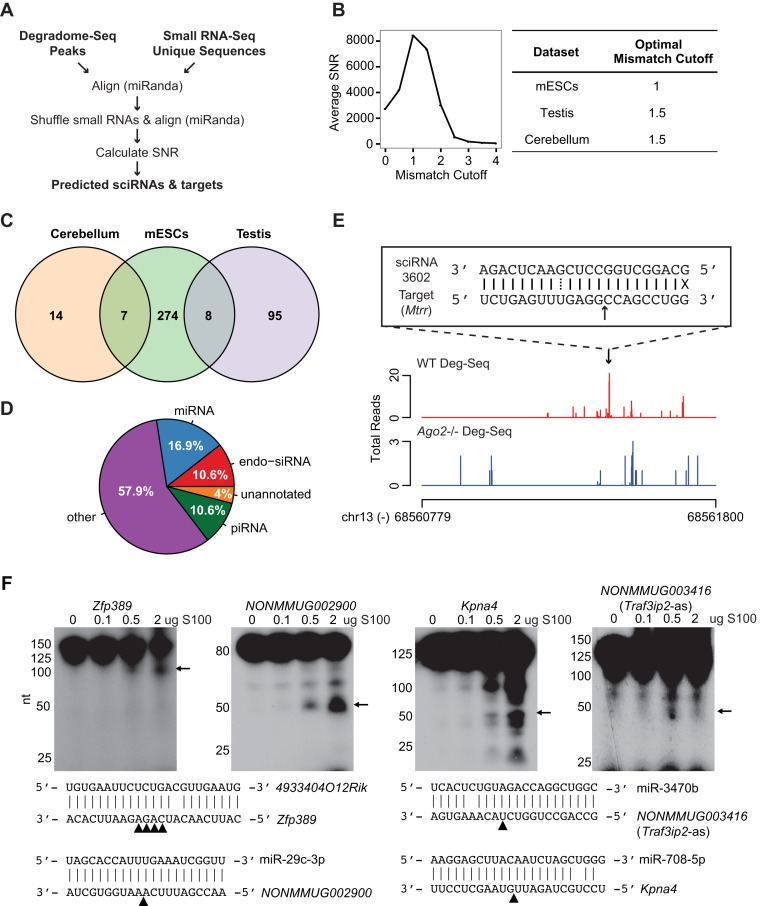
Figure 1.
Prediction of sciRNA-mediated mRNA cleavage events. (A) Bioinformatic pipeline schematic. (B) Left panel: average SNR for each mismatch cutoff in mESCs; right panel: optimal mismatch cutoff corresponding to the maximum average SNR for each data set. (C) Venn diagram of sciRNAs identified in mESCs, testis 6M PN, and cerebellum 6M PN. (D) Pie chart of sciRNA annotations (total = 398, combining sciRNAs from 3 cell types). (E) An example of predicted sciRNA-mediated cleavage. Read distributions are shown for the 3′ UTR of the Mtrr gene in Deg-Seq data of wild type (WT, red) and Ago2 knockout (blue) mESCs. Alignment of the sciRNA to the Deg-Seq peak is shown in a box, where a solid line indicates a base pair match, dotted line indicates a G = U wobble, X indicates a mismatch, and black arrow indicates location of the Deg-Seq peak. (F) Experimental validation of target RNA cleavage mediated by small RNAs. A total of 200 ng of Kpna4, NONMMUG002900, Zfp389 or NONMMUG003416 (Trafip2-as) RNA were incubated with different amount of HeLa S100 loaded with 50 nM synthetic sciRNA at 37°C for 30 min. Arrows indicate cleaved 5′ RNA fragments (whose sizes are consistent with predicted cleavage products). Small RNA/target RNA sequences are shown with arrowheads indicating the predicted cleavage sites (4 sites identified as Deg-Seq peaks in Zfp389).