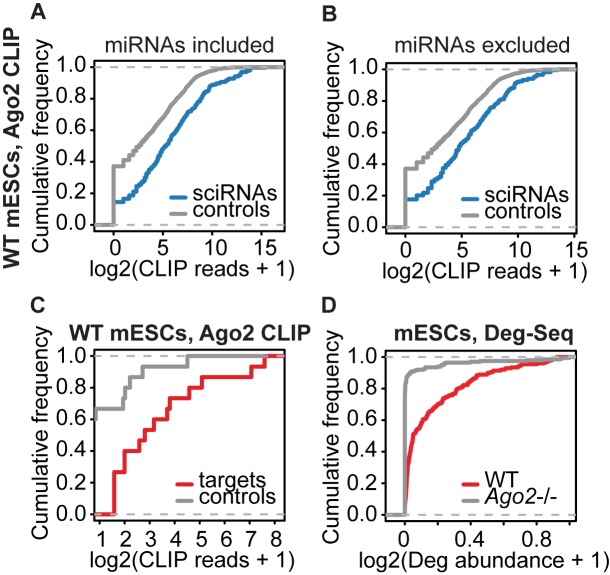
Figure 2.
Genomic data supporting the validity of sciRNA-target predictions. (A) Empirical cumulative frequency of abundance of Ago2 CLIP-Seq reads containing sciRNA sequences (blue) or control sequences randomly picked from Dicer-independent Dgcr8-independent small RNAs (grey) in WT mESCs (P = 2.2e-16, two-sided Kolmogorov–Smirnov (KS) test, same below). (B) Similar to (A), excluding miRNAs (P = 2.1e-10). (C) Similar to (A), comparing abundance of CLIP reads covering Deg-Seq peaks in target genes (red) or controls (grey, see Supplementary Methods) (P = 0.003). (D) Deg-Seq peak abundance in wild type (WT) and Ago2 knockout mESCs (P < 2.2e-16).