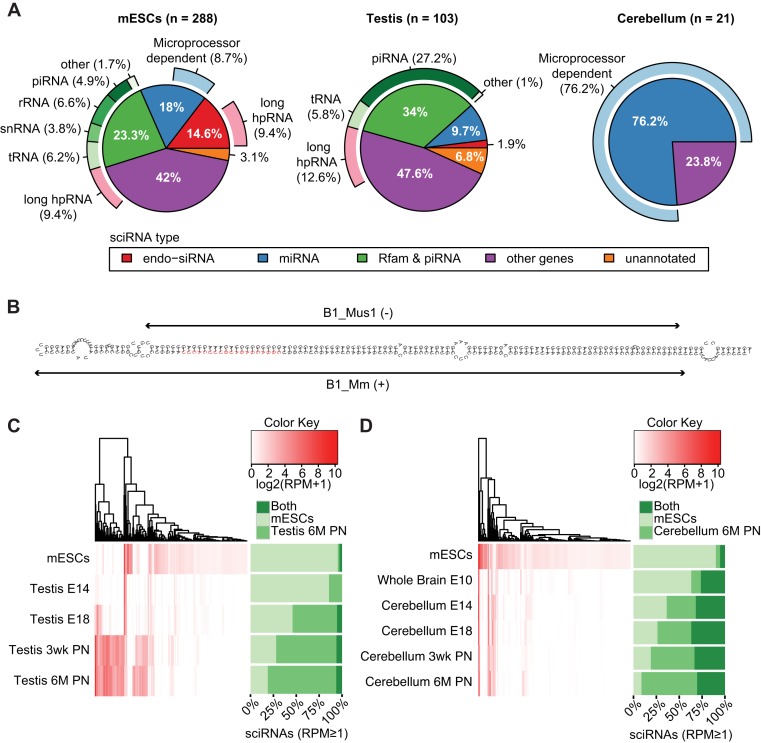
Figure 3.
Characterization of sciRNAs. (A) Categorization of sciRNAs in mESCs, testis and cerebellum according to: (i) annotation (inner circle), (ii) dependence on the microprocessor (outer circle, light blue) and (iii) long hpRNA structure (outer circle, pink). In mESCs, 1 unmapped sciRNA was excluded. (B) RNAfold structure of sciRNA 24792 (red) and flanking regions (within the Ccdc30 gene). This sciRNA was predicted in mESCs. Two inverted B1 repeats (Repeatmasker) are labeled. (C) Hierarchical clustering of sciRNA expression levels (reads per million, RPM) in mESCs and different stages of testis development. E14: embryonic day 14; E18: embryonomic day 18; 3wk PN: 3-week postnatal; 6M PN: 6-month postnatal. In the heatmap, RPM values of all sciRNAs that were identified originally in mESCs or 6M PN testis were visualized for each sample. Stacked bars on the right show the percentage of sciRNAs (among those with RPM ≥ 1) specific to mESCs (defined as those that were only identified in mESCs by the pipeline in Figure 1A, but not in the testis 6M PN data), testis 6M PN (similarly as defined above) or common to both. Note that some sciRNAs predicted originally in mESCs or testis may be excluded in the stacked bars due to low RPM. (D) Similar to (C), for cerebellum development. E10: embryonic day 10.