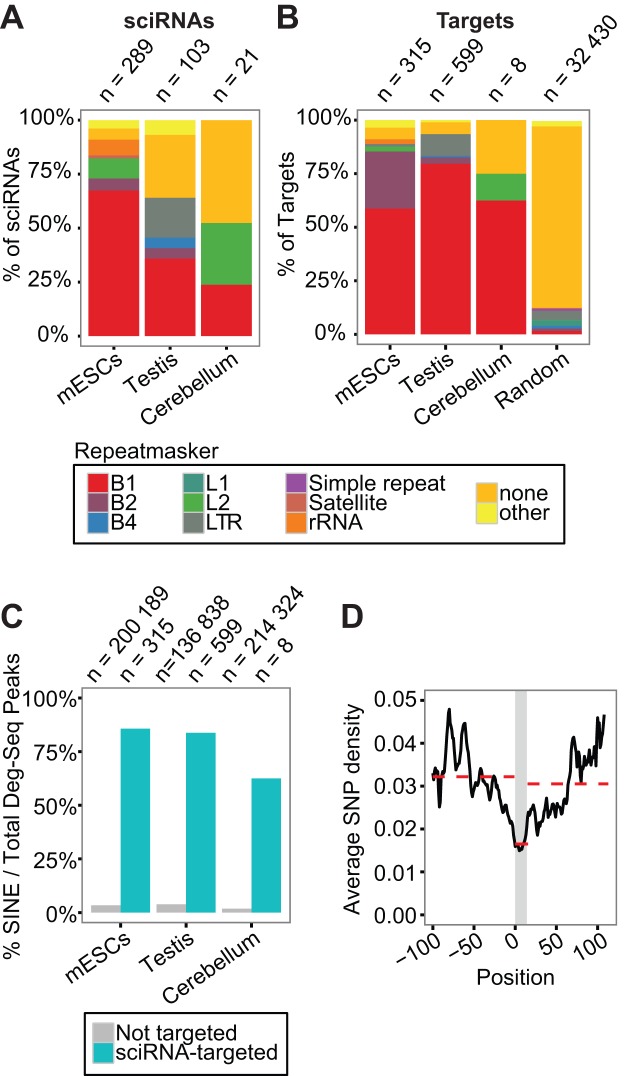
Figure 5.
Small RNA guided endonucleolytic cleavage targets retrotransposons. (A) Distribution of sciRNAs in different types of repeats (Repeatmasker). If more than one Repeatmasker annotation was identified, the following prioritization was used: B1 > B2 > B4 > others. Random: similar to Figure 4B. (B) Similar to (A), for target cleavage sites. (C) Percentage of Deg-Seq peaks overlapping SINE regions among all Deg-Seq peaks. Two groups of Deg-Seq peaks are shown: those targeted by sciRNAs (sciRNA-targeted) or otherwise (Not targeted). (D) Average SNP density per position in all B1 sequences bound by sciRNAs and their ±100 nt flanking region (chi-square P-value P < 2.2e-16). The y-axis is the average SNP density per nucleotide (see Supplementary Methods). A smoothing window of 10nt was applied to all data points. The grey region indicates the sciRNA-binding region. It ranges from 0–14 because the maximum length of targeted region was 24 and the smoothing window spanned 10 nt. Red dashed lines indicate the average SNP density of the three corresponding regions.