Figure 1. Phylogenetic analysis of H10 (A) and N8 (B) genes recovered from the samples collected from the LPM which the index patient visited.
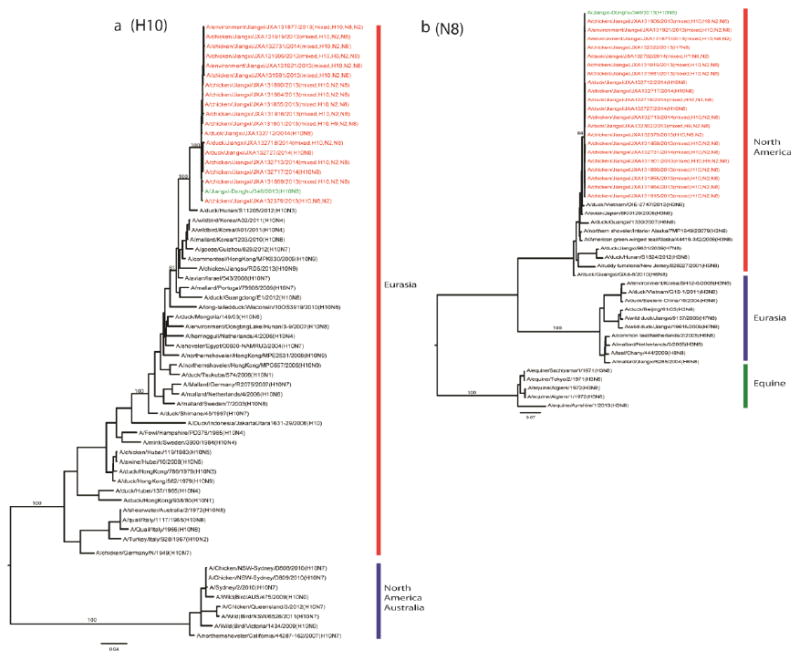
The phylogenetic analyses were performed using maximum likelihood by GARLI (Gutell and Jansen, 2006) and bootstrap resampling analyses using PAUP* 4.0 Beta (Wilgenbusch and Swofford, 2003) to apply a neighborhood joining method, as described earlier (Wan et al., 2008). The genes from this surveillance are marked in red, and those from the human H10N8 isolate in green. The bootstrap values for representative lineages are marked.
