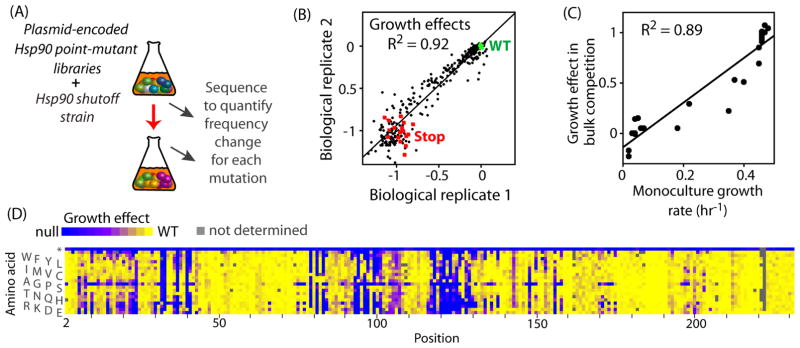
Figure 1. Effects of individual amino acid changes in the N-domain of Hsp90 on yeast growth rate.
(A) Outline of the bulk competition strategy utilized to analyze the effects of Hsp90 mutants on yeast growth rate. (B) Full biological replicates of Hsp90 mutations spanning amino acids 12-21 and 92-101 are strongly correlated. Growth effects are plotted as selection coefficients (0=no effect compared to wildtype, and −1=null). Silent mutations that do not change the protein sequence are plotted in green, stop codons are shown in red and amino acid changes in black. (C) Estimates of growth effects from bulk competitions correlate strongly with measurements of mono-culture growth rate for a panel of Hsp90 mutants. The following mutations were analyzed in monoculture: L18F, Y24L, R32A, N37S, D40E, K54E, E57K, R65L, Q72D, G83T, I96A, S99A, I117F, F120A, G121A, F124A, Y125W, L129M, E165K, I172D, and K191F. (D) Heatmap representation of the fitness landscape observed for individual amino acid changes across amino acids 2-231 of Hsp90. See also Figure S1 and tableS1.