FIGURE 2.
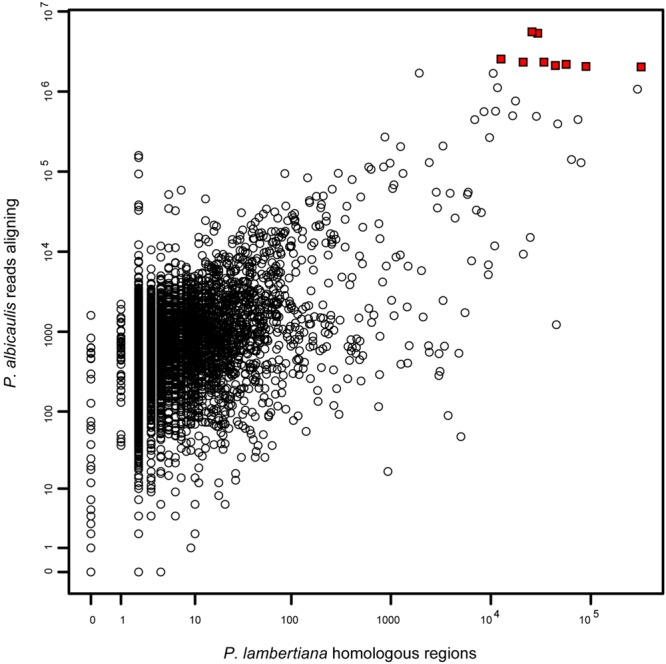
Coverage in Pinus albicaulis (aligned reads among all 48 samples) per transcript (median = 735), as a function of homologous regions in the P. lambertiana genome (median = 2) (log-log plot). Representation of transcript sequences was correlated among datasets, indicating that probes matching a large number of P. albicaulis reads are likely highly repetitive in the genome. The nine transcripts indicated by red squares all captured over two million reads each and together represent >50% of all aligning reads. The cause of this overrepresentation is that these regions are among the top 0.25% most repetitive regions in the genome, all with over 10,000 homologous regions found. Removal of a small number of highly repetitive probes in future studies would substantially increase the coverage of all other loci.
