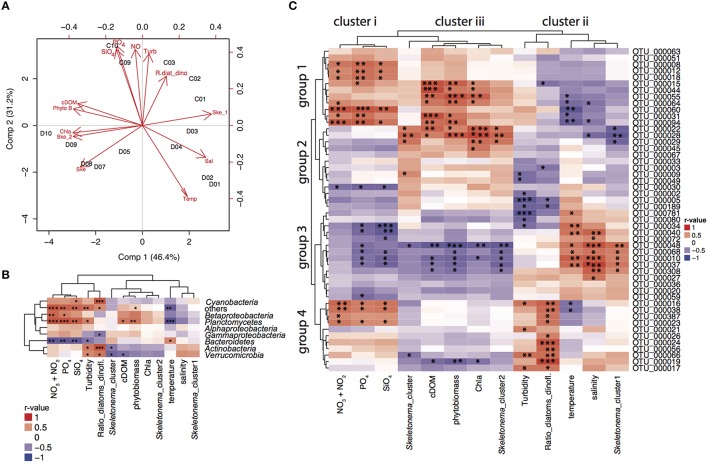
Figure 3.
PLSr and correlations to environmental variables. (A) Partial least squares regression analysis (PLSr) linking bacterial community composition (including all OTUs) with environmental variables and biotic factors in stations 1–3, 9–10 during cruise C, and 1–10 during cruise D. Red short labels describe the environmental variables (PO4; phosphate, NO; nitrate and nitrite, SiO4; silicate, Turb; turbidity, R.diat_dino; diatom/dinoflagelate ratio, cDOM; colored fluorescent DOM, Phyto B; phytoplankton biomass, Chla; chlorophyll a fluorescence, Ske_2; S. marinoi cluster 2, Ske; S. marinoi cluster, Temp; temperature, Sal; salinity, Ske_1; S. marinoi cluster 1) whereas black labels describe the sampling stations. (B) Correlation analysis between relative abundances of bacterial families and environmental variables based on Pearson correlations for relative abundances of bacterial families and environmental variables. Correlation values depict r-values of Pearson correlations and the dendograms are based on Pearson correlations. Statistical significance levels: *p < 0.05, **p < 0.01, ***p < 0.001. (C) Correlation analysis between relative abundances of 50 most abundant OTUs and environmental variables based on Pearson correlations. Correlation values depict r-values of Pearson correlations. Statistical significance levels: *p < 0.05, **p < 0.01, ***p < 0.001.