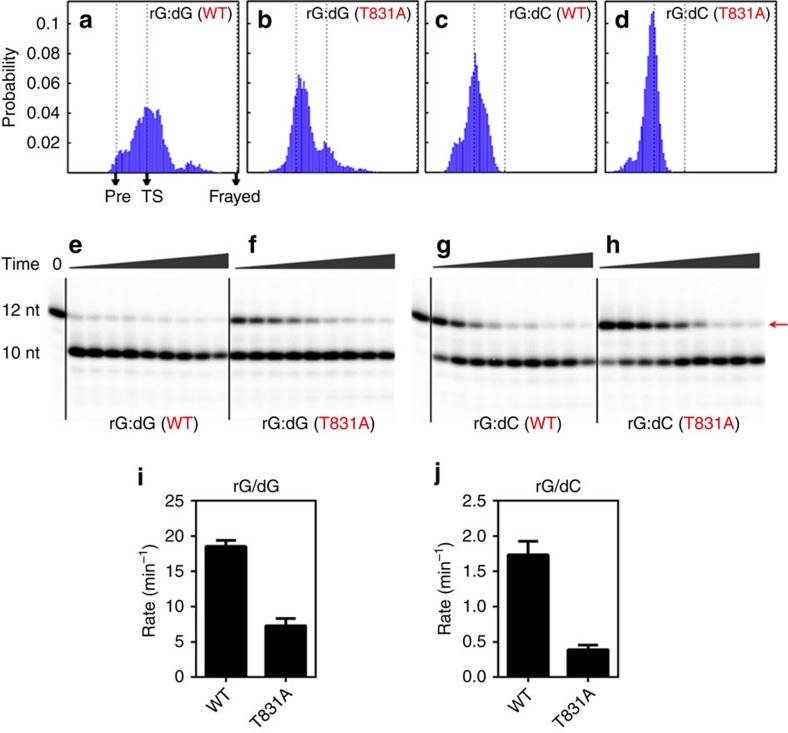
Figure 3. Fraying motion of the RNA 3′-end nucleotide is dictated by the mismatched base pair in the active site and promoted by BH residue T831 serving as a sensing probe.
As a comparison, one additional matched base pair (rG:dC) was designed for the mutant MD simulations starting from the regions near to the transition state (TS) between S1 and S2 states. (a–d) Histogram for the movements of the RNA 3′-end nucleotide for the rG:dG (a) and rG:dC (c) base pairs starting from the above TS, and their corresponding T831A mutants on the Pol II BH domain (b and d, respectively). Three critical states: Pre-translocation state (S1), TS and Frayed state (S2), are mapped with dashed lines and black arrows in the plots. The x coordinate represents a transition index, defined as 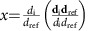 , where di=rMD−rpre is the vector connecting the pre-translocation structure and a certain MD conformation (each conformation r is represented using the c.o.m coordinates of its DNA TN). dref=rfrayed−rpre is the reference vector connecting the pre-translocation and frayed state. The transition index indicates where a MD conformation locates between the pre-translocation and frayed state. In particular, x=0 corresponds to the pre-translocation (pre) state while x=1 corresponds to the frayed state. (e–h) Evaluation of the functional roles of the BH residue T831 in backtracking by site-directed mutagenesis studies and TFIIS cleavage assays. Comparing to the WTs of the rG:dG and rG:dC systems (e and g, respectively), the corresponding T831A mutant has weaker TFIIS-facilitated cleavage activities (f and h, respectively). The upper bands refer to the initial RNA transcript (12 nt) as indicated by red arrow; the lower bands refer to the cleaved product (10 nt). Quantitative analysis of TFIIS-stimulated cleavage rates is shown in i and j. Error bars represent standard deviations derived from three independent experiments.
, where di=rMD−rpre is the vector connecting the pre-translocation structure and a certain MD conformation (each conformation r is represented using the c.o.m coordinates of its DNA TN). dref=rfrayed−rpre is the reference vector connecting the pre-translocation and frayed state. The transition index indicates where a MD conformation locates between the pre-translocation and frayed state. In particular, x=0 corresponds to the pre-translocation (pre) state while x=1 corresponds to the frayed state. (e–h) Evaluation of the functional roles of the BH residue T831 in backtracking by site-directed mutagenesis studies and TFIIS cleavage assays. Comparing to the WTs of the rG:dG and rG:dC systems (e and g, respectively), the corresponding T831A mutant has weaker TFIIS-facilitated cleavage activities (f and h, respectively). The upper bands refer to the initial RNA transcript (12 nt) as indicated by red arrow; the lower bands refer to the cleaved product (10 nt). Quantitative analysis of TFIIS-stimulated cleavage rates is shown in i and j. Error bars represent standard deviations derived from three independent experiments.