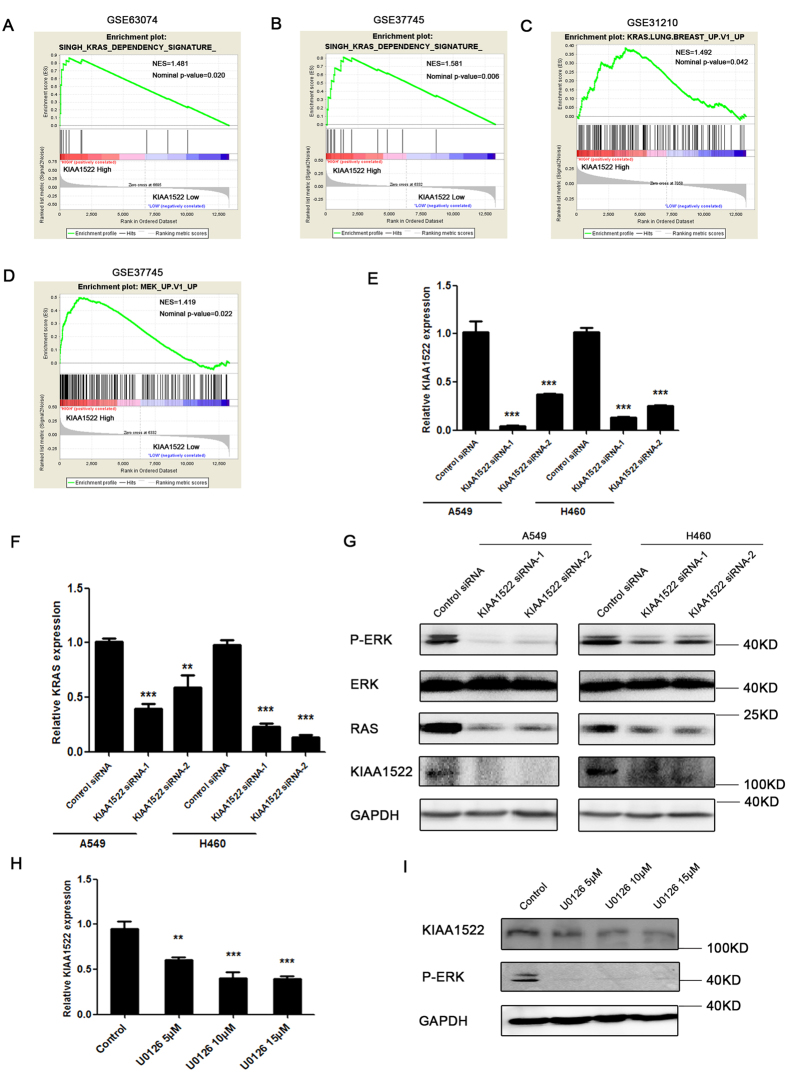
Figure 6. Correlation of KIAA1522 expression with KRAS pathway signatures.
(A–C) Gene sets enrichment analysis of the NSCLC dataset showing that high KIAA1522 expression was associated with the hyper-activation of KRAS signatures in dataset GSE63074 (A), GSE37745 (B) and GSE 31210 (C). (D) KIAA1522 high expression showed positive correlation with activation of MEK signaling in GSE37745. (E) The expression of KIAA1522 in the A549 and H460 cells that were transfected with control siRNA or two independent KIAA1522 siRNAs for three days. Data are shown as mean ± s.d. (by t-test analysis, *** P < 0.001, n = 3). (F) The relative KRAS mRNA levels in the A549 and H460 cells transfected with control siRNA or two independent KIAA1522 siRNAs for three days were analyzed by real-time PCR. Data shown are mean ± s.d. (by t-test analysis, ***P < 0.001, **P < 0.01, n = 3). (G) Western blotting analysis of the Phospho-ERK (Thr202/Tyr204), ERK, RAS, KIAA1522 and GAPDH (loading control) levels in the A549 and H460 cells transfected with the indicated siRNAs, four days after transfection. (H–I) The H460 cells were treated with DMSO (control) or the indicated concentration of U0126 for 24 hours; the cells were then subjected for real-time PCR (H) and western blotting (I) analysis of KIAA1522 expression. The efficiency of MEK inhibitor U0126 were revealed by the reduced Phospho-ERK (Thr202/Tyr204) levels tested by western blot. Data shown are mean ± s.d. (by t-test analysis, **P < 0.01, ***P < 0.001, n = 3).