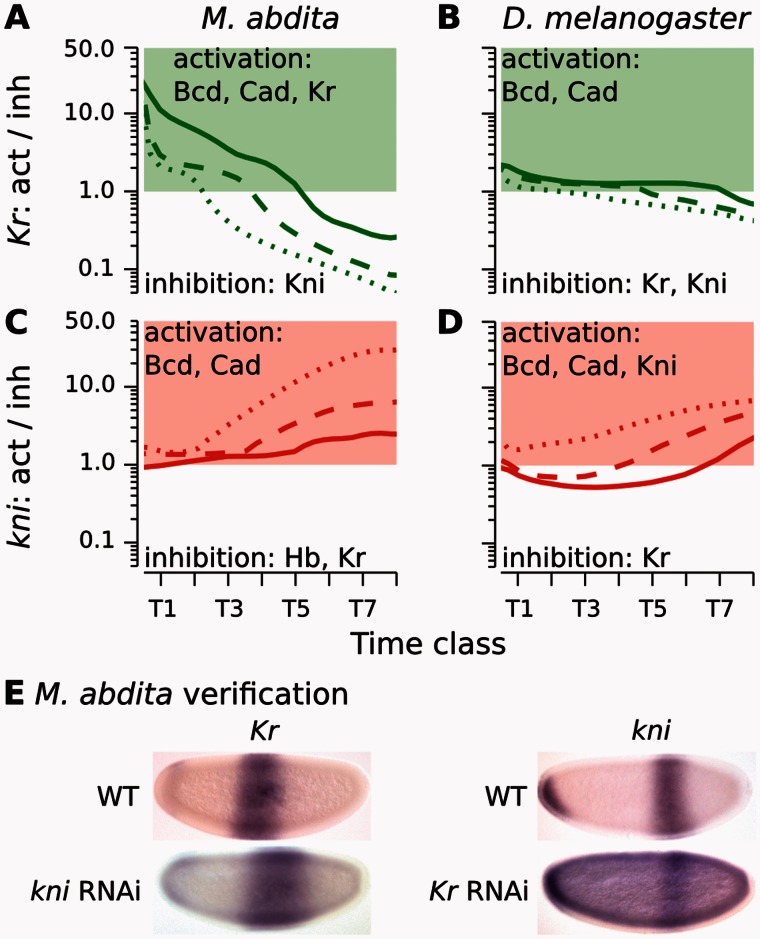
Fig. 4.
Graphical analysis of regulatory interactions involved in positioning the Kr–kni boundary interface. (A–D) Plots show ratios of activating versus repressive regulatory input on Kr (A, B) and kni (C, D) over time in Megaselia abdita (A, C) and Drosophila melanogaster (B, D). Lines indicate equidistant nuclei at 53% (solid), 55% (dashed), and 59% (dotted) A–P position (A, C), and 54% (solid), 56% (dashed), and 58% (dotted) A–P position, respectively (B, D). Green/red colored areas indicate activation of Kr/kni (>1.0), white areas indicate inhibition (<1.0). Despite subtle differences in shift mechanism and dynamics, both M. abdita and D. melanogaster show increasing Kr repression and kni activation over time due to repressive asymmetry between the two genes. See main text for details. (E) Embryos of wild-type (WT) and RNAi-treated M. abdita embryos stained for Kr and kni mRNA at time class T3. Embryos are shown in lateral view: Anterior is to the left, dorsal is up. Embryos represent illustrative examples drawn from a quantitative and systematic data set of M. abdita RNAi knockdowns (Wotton, Jiménez-Guri, Crombach, Janssens, et al. 2015).