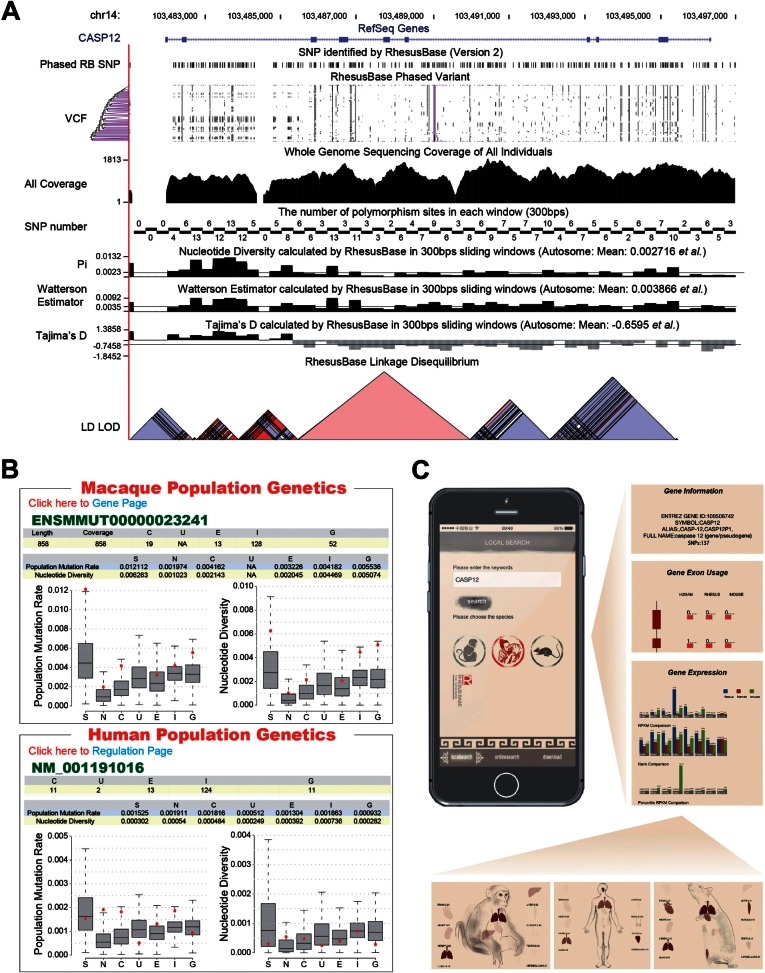
Fig. 2.
Evolutionary and functional interrogation of single genes in RhesusBase PopGateway. (A) An expanded view of RhesusBase Genome Browser at the CASP12 locus, shown with comprehensive position-based annotations, such as the gene structure, polymorphisms, nucleotide diversity (π), population mutation rate (θw), Tajima’s D, and linkage disequilibrium. (B) A typical RhesusBase Population Genetics Page showing the comparative population genetics features of CASP12 in human and rhesus macaque. (C) For each gene of interest, three annotation pages—the Gene Information, Gene Exon Usage, and Gene Expression—were developed for visualizing, respectively, the basic gene annotations, the exon splicing efficiency, and gene expression levels in a comparative view of human, rhesus macaque, and mouse. Detailed information could be further accessed by clicking the corresponding links. For example, the transcript expression of the gene of interest is shown in RPKM (Reads Per Kilobase of exon model per Million mapped reads) and marked on the body map, with the color intensity corresponding to its relative expression level in each tissue of the different species.