Figure 5.1. Chromatin remodeling complexes are segregated into distinct families.
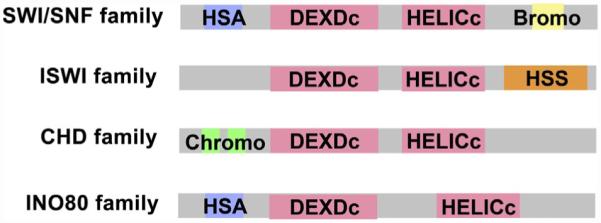
The ATPase containing subunits found in chromatin remodeling complexes have common functional domains allowing their segregation into distinct families. All ATPase subunits have distinctive DEXDc ATP-binding domains and a HELICc helicase domain. The combination of these domains convert the chemical energy stored in ATP into mechanical energy used to remodel nucleosomes. The space between these two domains varies among family members and is distinctively long for the INO80 family of ATPases. Other domains which are diagnostic to specific families include the HSA domain (HSA) found in SWI/SNF and INO80 families, bromodomains (bromo) found in SWI/SNF family members, chromodomains (chromo) found in the CHD family, and the HAND–SANT–SLIDE (HSS) cluster found in ISWI family members (Becker & Workman, 2013).
