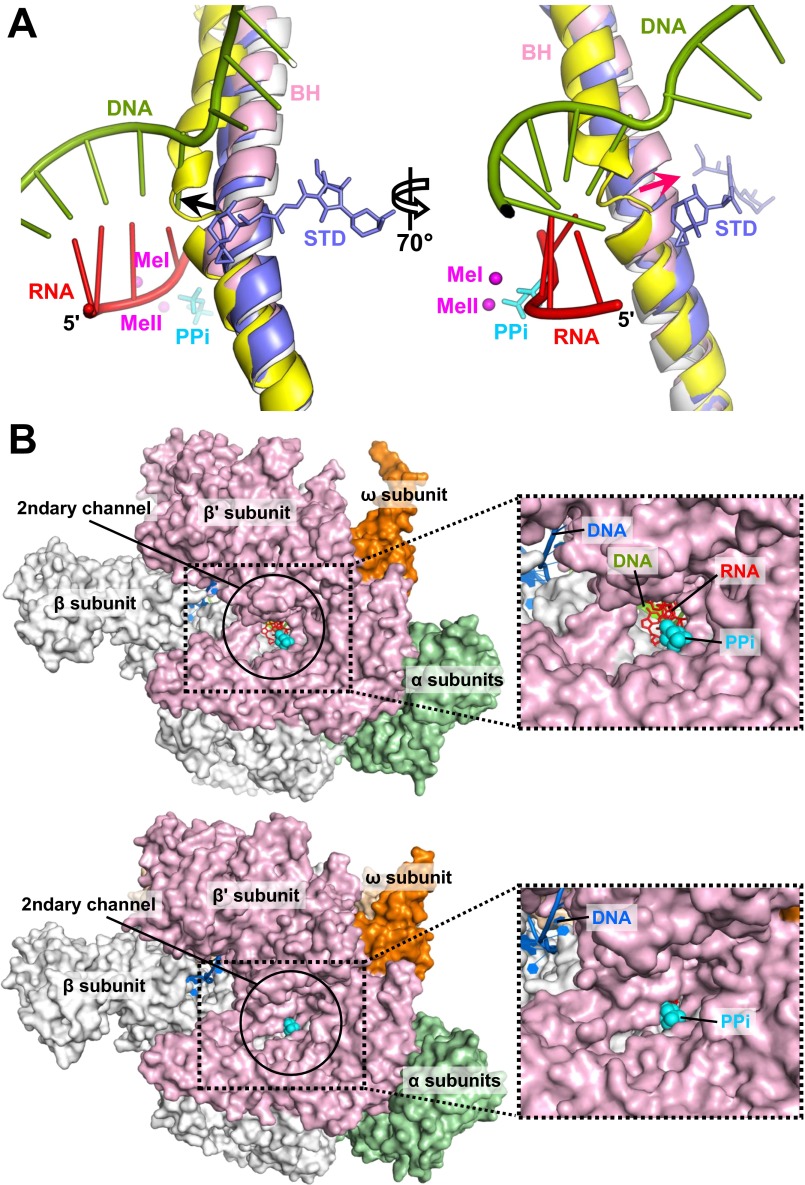
Fig. S4.
Active site conformations. (A) BH bending in the stressed TIC. Structures of the apo E. coli holoenzyme (PDB ID code 4LJZ) (gray), T. thermophilus elemental pause complex (PDB ID code 4GZY) (yellow), and T. thermophilus elongation complex with AMPcPP (APC) and streptolydigin (STD) (PDB ID code 2PPB) (blue) are superimposed using the RNAP β subunit. Streptolydigin and PPi (cyan) are presented as sticks. The nascent RNA and the T-strand DNA in the σS-TIC are shown as ladders. The black arrow marks the direction of BH bending related to transcription translocation; the red arrow marks the direction of BH bending observed in the TICs. Both directions of BH bending are based on a comparison with the RNAP apo holoenzyme. (B, Upper) An open active site. A view down to the secondary channel of the RNAP apo holoenzyme (PDB ID code 4LJZ) is presented. Nucleic acids and a PPi were modeled by superimposing the σS-TIC structure onto the RNAP apo holoenzyme using the β subunit. (Lower) A closed active site. A view down to the secondary channel of the σS-TIC (before NTP soaking) is displayed. The insets on the right are zoomed-in views of the secondary channel.