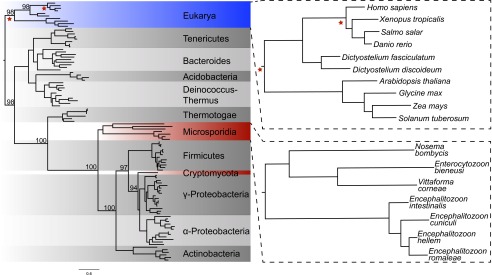
Fig. 3.
TK has been transferred from bacteria into Microsporidia and Cryptomycota by independent HGT events. Top BLASTP hits for E. cuniculi, No. ceranae, and R. allomycis TK were aligned along with TK sequences from representative eukaryotes and other bacterial phyla, and the resulting alignment was used to infer a maximum-likelihood phylogeny. Red stars indicate the location on the tree where likelihood-ratio tests were performed to evaluate the relationship of the sequences in question with Animalia or other Eukarya representatives. Bootstrap values for key nodes are noted. The N. parisii TK results from a third independent HGT, but is absent from the phylogenetic tree because of its distinctive viral domain structure and amino acid sequence (Fig. S2). Note that the No. ceranae TK has also been excluded due its long branch length (Dataset S2), but Nosema bombycis is shown.