Significance
With the spectacular advance of DNA-sequencing technology, a rapidly increasing number of subtle gene variants in the human population are being identified. One approach to understanding their impact on health and disease is to re-create these variants in appropriate cell lines that are accessible to functional analyses. We present a design principle for synthetic short single-stranded DNA oligonucleotides (ssODNs) that enables effective gene modification in DNA mismatch repair (MMR)-proficient cells. We report that the placement of locked nucleic acids at mismatching bases evades MMR. Its precision, simplicity, and cost-effectiveness makes ssODN-directed gene modification an attractive way of generating large numbers of sequence variants, in particular when they can be evaluated by scoring an easily detectable phenotype.
Keywords: subtle gene modification, DNA mismatch repair, locked nucleic acid, single-stranded oligonucleotides, embryonic stem cells
Abstract
Synthetic single-stranded DNA oligonucleotides (ssODNs) can be used to generate subtle genetic modifications in eukaryotic and prokaryotic cells without the requirement for prior generation of DNA double-stranded breaks. However, DNA mismatch repair (MMR) suppresses the efficiency of gene modification by >100-fold. Here we present a commercially available ssODN design that evades MMR and enables subtle gene modification in MMR-proficient cells. The presence of locked nucleic acids (LNAs) in the ssODNs at mismatching bases, or also at directly adjacent bases, allowed 1-, 2-, or 3-bp substitutions in MMR-proficient mouse embryonic stem cells as effectively as in MMR-deficient cells. Additionally, in MMR-proficient Escherichia coli, LNA modification of the ssODNs enabled effective single-base-pair substitution. In vitro, LNA modification of mismatches precluded binding of purified E. coli MMR protein MutS. These findings make ssODN-directed gene modification particularly well suited for applications that require the evaluation of a large number of sequence variants with an easy selectable phenotype.
Since the rapid decline in genome sequencing costs and the accumulation of data from genome-wide association studies, thousands of single-nucleotide variations in disease-related genes have been identified. One approach to functionally investigate these variants of uncertain clinical significance is to introduce them into the endogenous gene of appropriate model systems that are readily accessible to phenotypic assessment. Recently developed protocols for subtle gene modification are based on template-directed repair of DNA double-strand breaks (DSBs) that are site-specifically introduced by zinc-finger, TALE, or RNA-directed Cas9 nucleases (1). In particular, subtle mutations can effectively be introduced by single-stranded DNA oligonucleotides (ssODNs) carrying the modification of interest that serve as templates for the repair of CRISPR/Cas9-introduced DSBs (2). Several laboratories, including ours, have previously shown that ssODNs can also be used for subtle gene modification in the absence of DSBs. Although less efficient than nuclease-assisted gene modification, oligonucleotide-directed gene modification (also referred to as “oligo targeting”) is attractive due to its lack of additional components, simplicity, and cost-effectiveness, especially in cases where the consequences of the planned modifications can be scored by a selectable or easily detectable phenotype.
Different models have been proposed for the mechanism of oligo targeting (reviewed in ref. 3). Substantial evidence has been obtained that gene modification takes place during DNA replication (4–7). In this model, the ssODN anneals to single-stranded DNA in the replication fork, where it can serve as a primer for DNA synthesis by replicative polymerases (8). This process thus physically incorporates the ssODN and delivers the mutation to only one of the DNA strands (7, 9). Consistent with this view, thymidine and hydroxyurea—compounds that reduce the speed of replication—had a positive effect on targeting efficiencies, whereas replication arrest suppressed the targeting efficiency (10, 11).
In Escherichia coli, oligo targeting is strongly promoted by the bacteriophage λ-red–mediated recombination system (12–14). The phage-encoded Beta protein can bind oligonucleotides >35 nucleotides (nt) in length, thereby providing protection from degradation and promoting annealing to complementary single-stranded DNA (15–19).
In mammalian, yeast, and prokaryotic cell systems, it has unequivocally been demonstrated that the DNA mismatch repair (MMR) system has a strong suppressive effect on oligo targeting efficiencies (14, 20, 21). Disabling the MMR system through disruption of the key MMR gene Msh2 increased the targeting efficiency up to 700-fold in mouse embryonic stem cells (mESCs) (20). In an E. coli mutS-deficient strain, oligo targeting was up to 500-fold more effective than in MMR-proficient bacteria (14). Apparently, the mismatch that is created by annealing of the ssODN to its chromosomal complement creates a mismatch that is recognized by the MMR system. Mismatch recognition elicits a repair reaction that prevents incorporation of the oligonucleotide and aborts the gene modification reaction. Inconveniently, however, MMR deficiency results in the gradual accumulation of undesired mutations over time and may thus lead to secondary effects (22–26). Procedures that transiently suppress MMR—e.g., by using RNA interference (27, 28) or temperature-sensitive mutS and mutL mutants (29)—can create a short time window of MMR deficiency that enables effective subtle gene modification. Nevertheless, the accumulation of undefined mutations cannot be totally avoided and may confound the phenotype of the mutation under study (28).
An attractive solution to this problem is to avoid the action of MMR exclusively at the site of the targeted base substitution. Previous work in this direction has demonstrated that base analogs in ssODNs at the site of the substitutions can partially evade MMR in E. coli and in HeLa cells (30, 31). Thus far, the use of 2′-fluorouracil and 2-aminopurine yielded a twofold increased efficacy of substitutions to T and A, respectively, in MMR-proficient HeLa cells (31).
Here we present an ssODN design that allows complete evasion of MMR irrespective of the type of substitution. We demonstrate that inclusion of locked nucleic acids (LNAs) in the ssODNs at mismatching bases only, or also at directly adjacent bases, allows effective oligo targeting in MMR-proficient cells. LNA nucleotides contain a modified sugar residue with an additional 2′-C,4′-C-oxymethylene linker that confines the ribose ring to the 3′-endo conformation (32, 33). DNA:LNA duplexes demonstrate an increased stability and are more resistant to nucleases (34, 35). We report that LNA-modified ssODNs (LMOs) yielded equal efficiencies in MMR-proficient and -deficient mESCs for the substitution of one to three bases. For single- and four-nucleotide insertions in mESCs, we found partial evasion of MMR by LMO designs. We also analyzed the LMO design for generating single-nucleotide substitutions in E. coli and found that LMO efficiencies were independent of MMR status. Finally, we demonstrate that LNA modification renders a mismatch invisible to the MMR system.
Results
LNA Modification Allows Single-Base-Pair Substitution in MMR-Proficient Cells.
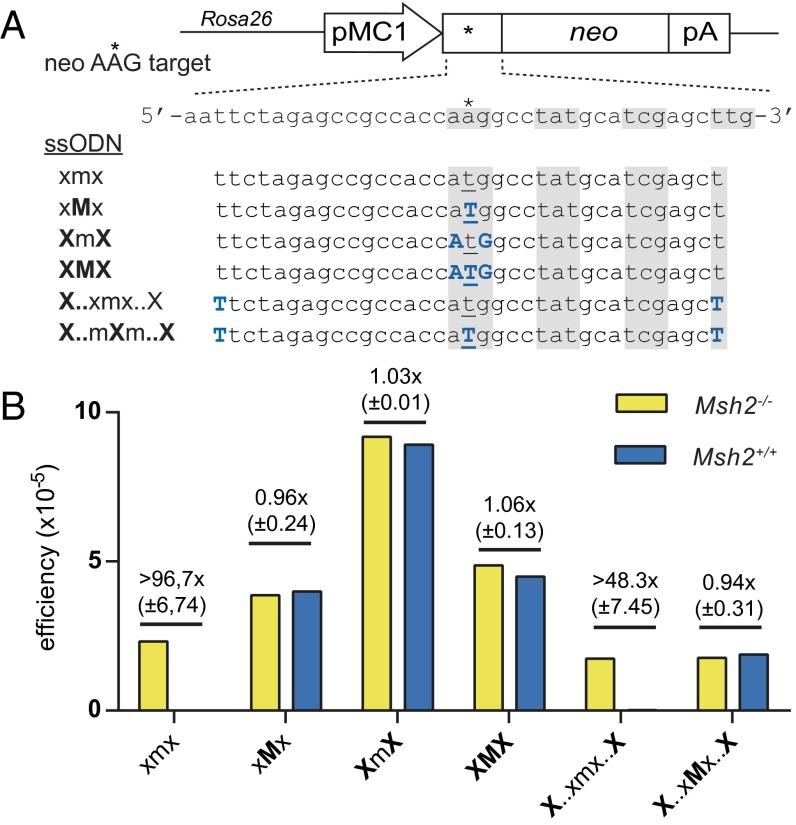
As readout for the efficiency of ssODN-directed gene modification, we made use of previously published MMR-deficient Msh2−/− and MMR-proficient Msh2+/+ mESCs that contain a single neomycin (neo) resistance gene, integrated into the Rosa26 locus, with a defective start codon, AAG (20). The efficiency of a 35-nt ssODN designed to generate a single A-to-T substitution that restores the start codon and thereby confers G418 resistance was strongly suppressed by DNA MMR (Fig. 1; ref. 20). Remarkably, we found that a single LNA modification at the mismatching nucleotide in the ssODN resulted in an efficiency of ∼5 × 10−5 in both Msh2−/− and Msh2+/+ cells (Fig. 1). The equal efficiencies in MMR-proficient and -deficient cells indicate that a single LNA modification was sufficient to overcome the suppressive effect of MMR. An LMO with LNAs flanking the mismatch evaded MMR as well and yielded an efficiency of ∼9 × 10−5. Three consecutive LNAs (at and flanking the mismatching base) also evaded MMR, whereas positioning LNAs at the 5′ and 3′ termini of the ssODN had no effect. We conclude that LNA modification at or in the direct vicinity of the mismatching nucleotide was required for MMR evasion.
Fig. 1.
Single base pair substituting LMOs evade MMR. (A) Schematic of the neo AAG reporter construct integrated in mESCs and sequences of LMOs used to generate a functional start codon by a single substitution. Asterisk indicates the T > A mutation in the defective start codon. Blue uppercase characters indicate LNA modifications. Alternating shaded areas highlight the introduced functional start codon and following codons; mismatching bases are underlined. (B) Targeting efficiency of LMOs expressed as the ratio of G418-resistant colonies over the number of seeded Msh2−/− or Msh2+/+ cells. The mean efficiency of two to four experiments is given, as well as the mean fold difference between Msh2−/− and Msh2+/+ cells with SD.
LMO Design for 2- and 3-bp Substitutions.
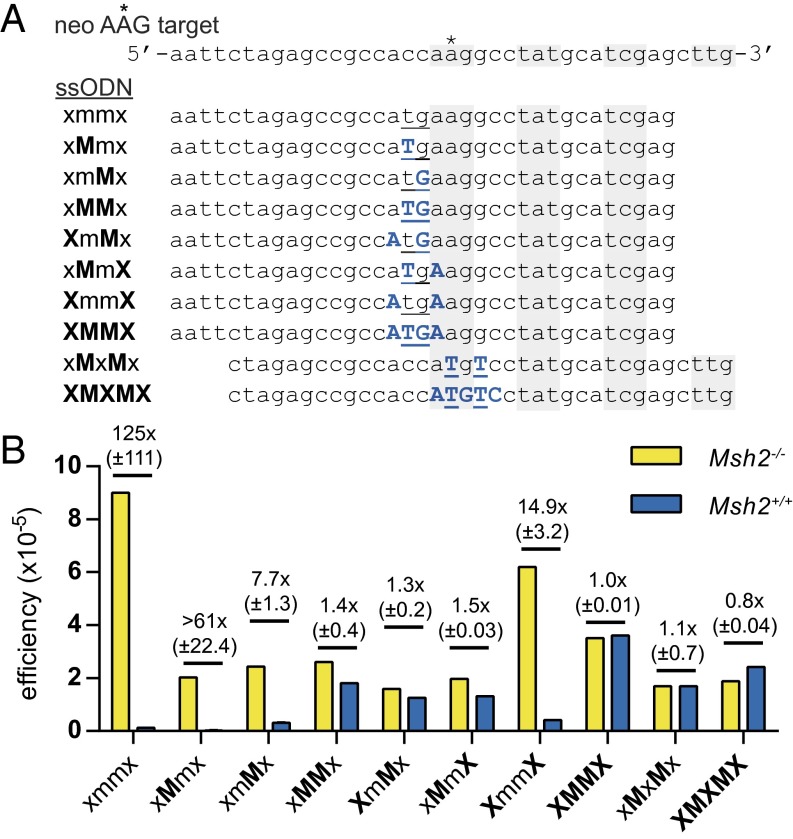
Next, we assessed the effectiveness of LMOs for multiple substitutions. We started with evaluating the efficiency of eight LMOs designed to generate a start codon by substituting two consecutive nucleotides and that contained one, two, or four LNAs at variable positions (Fig. 2A).
Fig. 2.
Evasion of MMR by LMOs introducing two substitutions. (A) neo AAG reporter sequence and sequences of LMOs used to generate a functional start codon by two base substitutions. Blue uppercase characters indicate LNA modifications; mismatching bases are underlined. Alternating shaded areas highlight the introduced functional start codon and following codons; mismatching bases are underlined. (B) Targeting efficiencies of the LMOs are expressed as the ratio of G418-resistant colonies over the number of seeded Msh2−/− or Msh2+/+ mESCs. The mean efficiency of two experiments is given, as well as the mean fold difference between Msh2−/− and Msh2+/+ cells with SD.
We found that for 2-bp substitutions in Msh2+/+ cells, two LNA modifications, of which at least one should be located at a mismatching base, were necessary to effectively evade MMR (Fig. 2B). An efficiency of 3.6 × 10−5 was reached with an LMO that contained four LNA modifications positioned at and adjacent to the mismatching bases. Furthermore, we designed two LMOs that were intended to generate two substitutions spaced by one base. One substitution restored the defective start codon, whereas the other replaced the first position of the second codon. Both LMOs, either containing two LNA nucleotides at the mismatching bases or five consecutive LNAs, were equally efficient in Msh2−/− and Msh2+/+ cells. Although these LMO designs successfully evaded MMR, we recognized that the efficiency reached was twofold to threefold reduced in comparison with the unmodified ssODN in Msh2−/− cells.
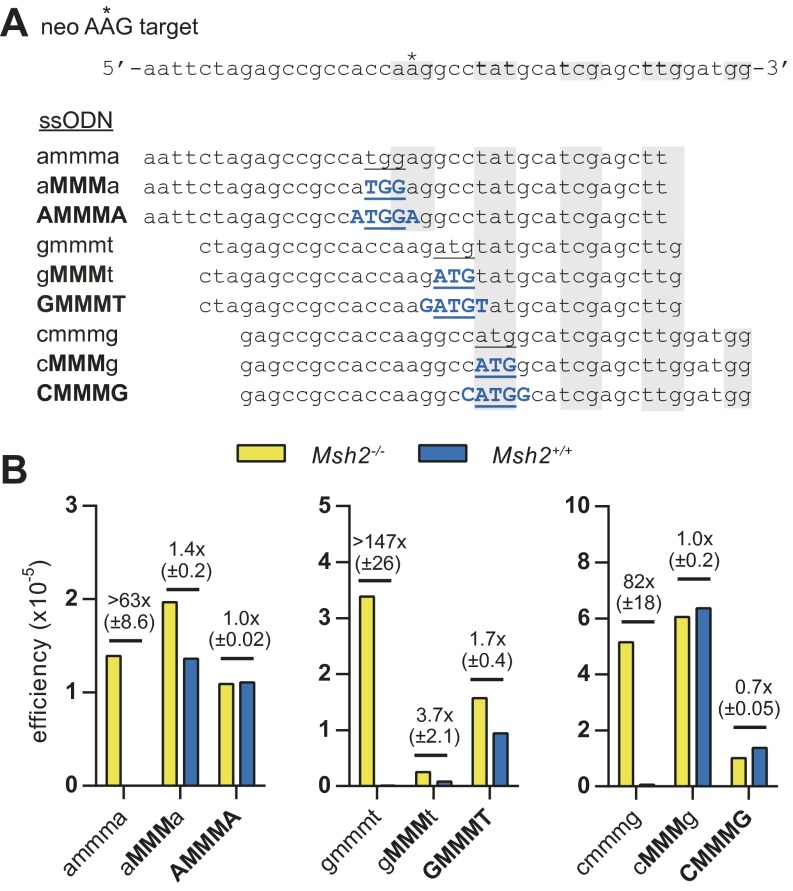
Finally, we explored the applicability of LMOs for the substitution of an entire codon in MMR-proficient cells. We designed three sets of LMOs, each generating a novel start codon by substituting three consecutive bases (Fig. S1). In two of the three codon substitutions made, we found that LNAs at the three mismatching bases led to equal efficiencies in Msh2−/− and Msh2+/+ cells. In one case, the presence of three consecutive LNA modifications was not sufficient, but for all three sets, we found that five LNAs placed at the three modifying bases plus the two adjacent bases completely evaded MMR (Fig. S1).
Fig. S1.
Evasion of MMR by LMOs generating a triple substitution at three different sites. (A) neo AAG reporter sequence and sequences of LMOs used to generate a functional start codon by three base substitutions. Blue uppercase characters indicate LNA modifications; mismatching bases are underlined. Alternating shaded areas highlight the introduced functional start codon and following codons; mismatching bases are underlined. (B) Targeting efficiencies of the LMOs are expressed as the ratio of G418-resistant colonies over the number of seeded Msh2−/− or Msh2+/+ mESCs. The mean efficiency of two experiments is given, as well as the mean fold difference between Msh2−/− and Msh2+/+ cells with SD.
We report that ssODNs containing strategically placed LNA nucleotides at mismatching bases enables single, double, and triple base-pair substitutions in MMR-proficient cells with equal efficiencies as in cells lacking a key MMR protein.
Partial Evasion of MMR by LMOs for Insertions of 1 and 4 bp.
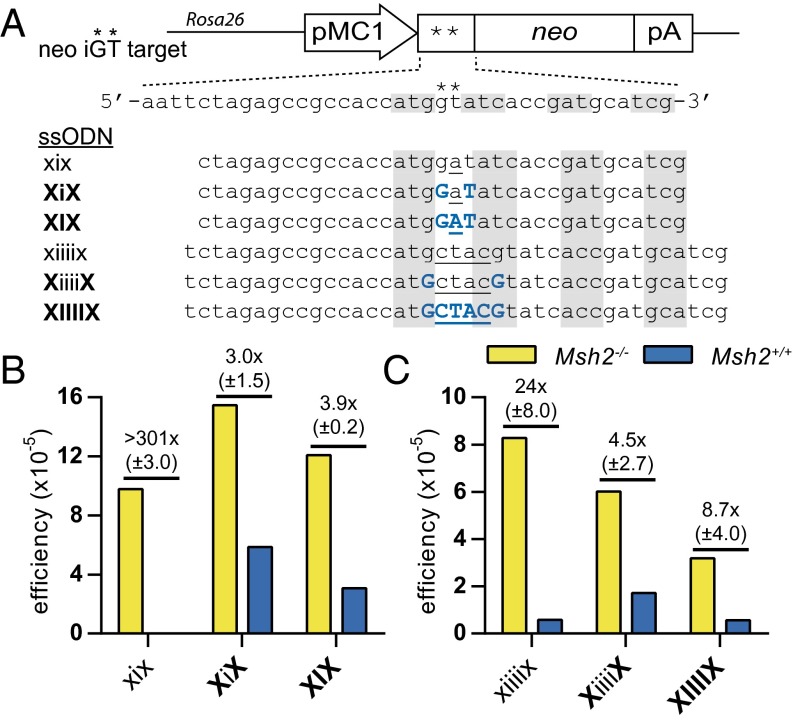
We have previously shown that nucleotide insertions can be achieved by ssODNs as well (20, 36). However, MMR needed to be abrogated because short loops of extrahelical nucleotides are subject to MMR. Loops of 1 and 2 nt are predominantly recognized by the MSH2–MSH6 heterodimer (37), whereas MSH2–MSH3 has a higher affinity for loops of 2–5 nt (38). Hence, the introduction of small insertions by ssODNs was suppressed in MMR-proficient cells (20, 36).
To test whether we could use LMOs for the generation of small insertions, we used a second neo reporter in which the start codon was rendered out of frame by a 2-bp GT insertion. Two sets of LMOs were designed that could restore the neo ORF by insertion of one or four bases, respectively. For the insertion of a single base pair, we found that placement of LNA nucleotides flanking the inserted base increased the frequency of gene modification in MMR-proficient cells 450-fold (Fig. 3B). The benefit was less pronounced when the inserted base also contained an LNA modification. As previously shown, an unmodified ssODN that inserted four bases was already effective at 0.6 × 10−5 in MMR-proficient cells, indicating that four extrahelical nucleotides were less well recognized by the MMR system. Nonetheless, we observed an approximately threefold efficiency increase for an LMO with LNA nucleotides flanking the insertion (Fig. 3C). Again, no further benefit was observed when the inserted nucleotides were LNA modified. Together, LMOs can be used for nucleotide insertions in MMR-proficient cells. However, they only partially evaded MMR as they were twofold to fourfold less effective than in Msh2−/− cells (Fig. 3).
Fig. 3.
Partial evasion of MMR by LMOs inserting one or four bases. (A) Schematic of the out-of-frame neo iGT reporter construct in mESCs and sequences of LMOs used to restore the neo ORF by 1- or 4-bp inserts. Asterisks indicate the inserted GT dinucleotide. Blue uppercase characters indicate LNA modifications; inserted bases are underlined. Alternating shaded areas highlight the start codon and following codons; mismatching bases are underlined. (B and C) Targeting efficiencies of single-nucleotide (B) and 4-nt (C) insertion LMOs are expressed as the ratio of G418-resistant colonies over the number of seeded Msh2−/− or Msh2+/+ mESCs. The mean efficiency of two experiments is given as well as the mean fold difference between Msh2−/− and Msh2+/+ cells with SD.
Improving Targeting Efficiencies in mESCs.
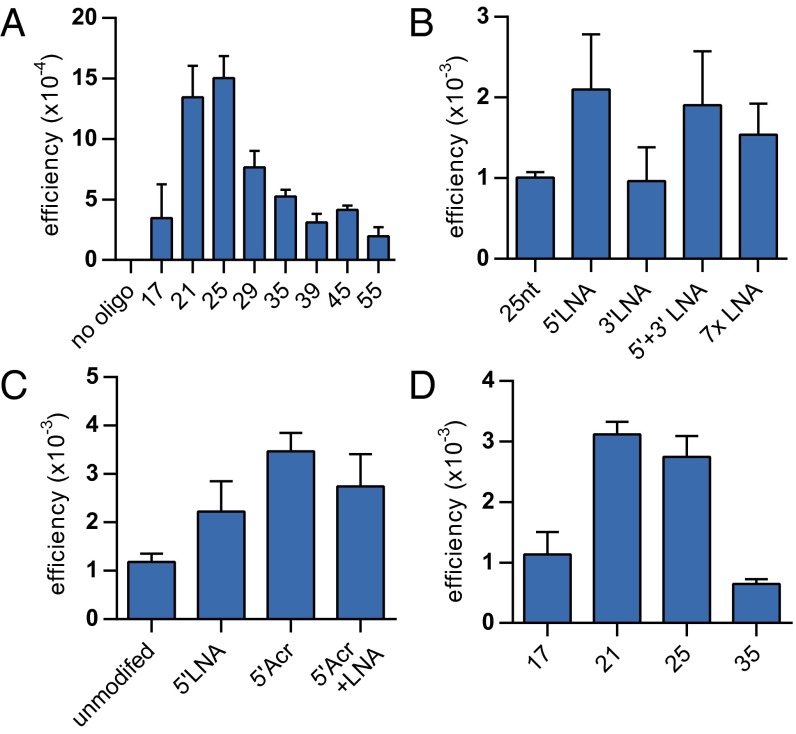
After establishing MMR evasion using LMOs, we evaluated whether these LMOs would benefit from further protocol optimization. First, we found that the use of an alternative transfection reagent, TransIT-Siquest (Mirus), increased targeting efficiencies approximately threefold. Second, we determined the optimal length of LMOs for single-nucleotide substitutions in Msh2+/+ cells. Although we previously found 35 nt to be optimal for unmodified ssODNs (11), we now found an optimum at 25 nt, reaching an efficiency of 1.5 × 10−3 (Fig. 4A). Third, because of the smaller optimal size of LMOs, we reasoned that protection from nucleolytic degradation might become increasingly relevant. Therefore, we tested LMOs with LNA-modified termini. Whereas terminal LNA modifications did not improve the efficacy of a 35-nt LMO (Fig. 1B), a single 5′ terminal LNA nucleotide added to a 25-nt LMO increased the efficiency twofold (Fig. 4B). By contrast, a 3′ LNA cap did not enhance the targeting efficiency. The addition of regularly spaced internal LNAs also did not further improve the targeting efficiency. Finally, we tested an LMO carrying a 9-amino-6-chloro-2-methoxy acridine moiety at its 5′ terminus, which may stabilize the annealing of ssODNs to the target site through DNA intercalation (39). This design was previously found to increase the targeting efficiency up to 65-fold (40). We found that the addition of a 5′ acridine to the 25-nt single-substitution LMO increased the targeting efficiency approximately threefold, resulting in an efficiency of 0.35% (Fig. 4C). Further downsizing the 5′ acridine LMO or combining it with a 5′ LNA did not significantly increase the efficiency (Fig. 4D).
Fig. 4.
Optimization of LMO design for A > T substitution in Msh2+/+ mESCs to restore neo AAG. All LMOs contain an LNA at the mismatching nucleotide. (A) Targeting efficiency of 300 pmol of LMOs ranging from 17 to 55 nt in size. (B) Modification of LMOs with 5′ and/or 3′ LNA termini or internal LNAs (7xLNA). (C) The 5′ terminus modification of LMOs with LNA and/or 9-amino-6-chloro-2-methoxy-acridine (Acr). (D) Transfection of 300 pmol of 5′Acr modified LMOs ranging from 17 to 35 nt in size. Data are from three experiments; error bars indicate SD. Transfection was prepared by using TransIT-siQUEST.
Sequence Analysis of LMO-Targeted mESC Clones.
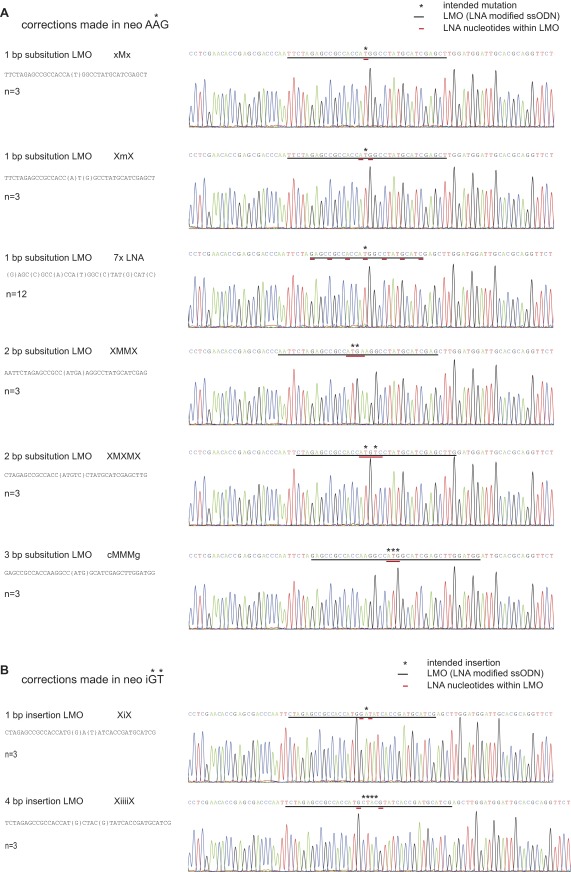
To determine whether LMOs induce inadvertent mutations in the vicinity of the planned substitution in MMR-proficient mESCs, we sequenced the genomic sequence from ±85 bp upstream to ±250 bp downstream of the neo start site of multiple G418-selected clones obtained with LMOs aimed to generate 1-, 2-, and 3-bp substitutions (Fig. S2A) and 1- and 4-bp insertions (Fig. S2B). In addition, we determined the sequence of 12 G418-resistant colonies that were generated with a 25-nt LMO containing seven LNA modifications (Fig. S2A). All 33 analyzed colonies contained the intended substitution or insertion. Moreover, none of the sequences showed additional variations. Thus, LMO targeting in MMR-proficient mESCs was highly precise.
Fig. S2.
Sequence analysis of neo start region in individual G418-resistant MMR-proficient mESC colonies. (A) Sanger sequencing results from G418-resistant colonies generated with LMOs designed to substitute 1, 2, or 3 bp. (B) Sanger sequencing results from G418-resistant colonies generated with LMOs designed to insert 1 or 4 bp. Black and red underscores highlight the LMO sequence and LNA modifications, respectively. Asterisks above sequence indicate the intended mutation.
LNA Modification Allows Base Pair Substitution in MMR-Proficient E. coli.
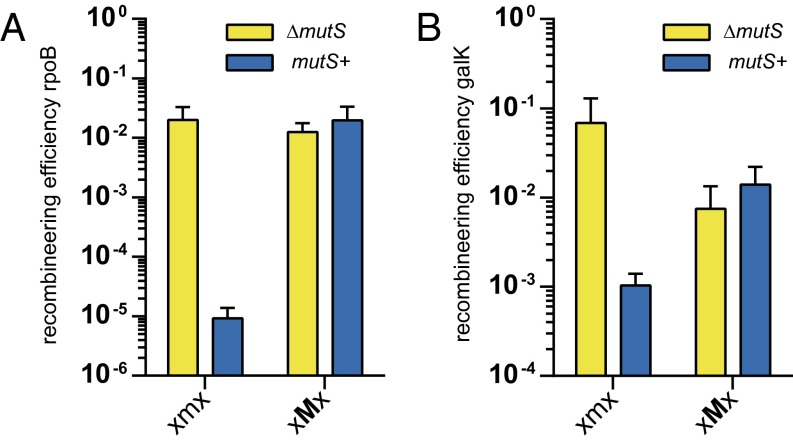
The principle of ssODN-directed gene modification has also been used to generate mutations in E. coli. A highly effective protocol has been developed that takes advantage of a bacteriophage λ-red–encoded recombination system (12, 13). However, also in E. coli, the efficiency appeared to be strongly repressed by MMR (14). To test the applicability of the LMO design in E. coli, we designed two sets of 70-nt LMOs that generate a single base pair substitution in either rpoB or galK. These LMOs contained a single LNA nucleotide at the position of the introduced mismatch. We electroporated these LMOs into published MMR-proficient (mutS+) and MMR-deficient (∆mutS) recombineering proficient E. coli strains (13, 14).
We aimed to generate mutation D516N (by mutating GAC to AAC) in rpoB, which results in rifampicin resistance (41). For galK, we aimed to correct the previously introduced Y145 stop codon, thereby enabling bacterial growth on minimal medium with galactose as the sole carbon source (13). At the rpoB locus, unmodified ssODNs and LMOs performed equally well in MMR-deficient cells, whereas in MMR-proficient cells, only the LMO efficiently introduced the D516N mutation (Fig. 5A). Also for galK, we found that the LMO outperformed the unmodified ssODN in MMR-proficient cells, even though the LMO was six times less efficient then the unmodified ssODN in the MMR-deficient strain (Fig. 5B). These results demonstrate that also in E. coli, ssODN-directed gene modification strongly benefits from LNA modification.
Fig. 5.
LMOs evade MMR in E. coli during λ-red–mediated recombineering. (A) Targeting efficiency of LMOs at the rpoB locus to generate D516N. (B) Targeting efficiency of LMOs correcting the GalKTyr145UAG stop codon. xmx indicates the mismatching base of the ssODN together with two adjacent bases; capital letters indicate LNA modifications. Data are from at least three (rpoB) or four (galK) independent electroporations; error bars indicate SD.
LNA Modification of Mismatches Prevents Binding of Bacterial MutS.
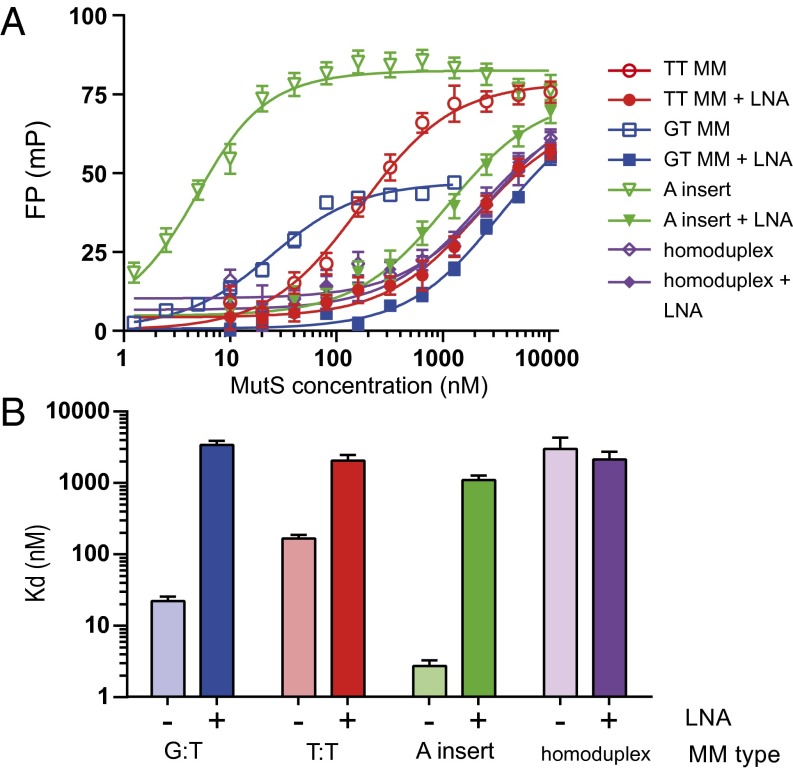
To explain why LMOs evade MMR, we tested whether LNA modification of mismatches interferes with mismatch recognition. To this aim, we measured equilibrium binding of purified bacterial MutS to LNA-modified mismatches by fluorescence polarization (FP). FP measurements were performed by using Cy5-labeled DNA containing either a GT or TT mismatch or an A insert. The various unmodified heteroduplexes were bound by MutS with different affinities, whereas MutS binding to homoduplex DNA was weak (Fig. 6 A and B). Strikingly, we found that a single LNA modification at the mismatch resulted in loss of binding, such that binding curves overlap with the homoduplex binding curve. The MutS binding constants for LNA-modified mismatches were similar to that of homoduplex DNA (Fig. 6B). In addition, for the DNA containing an A insert with two flanking LNAs, we found a dramatic reduction in MutS binding affinity. In the latter case, some weak affinity seemed to be maintained (Fig. 6B). This weak affinity is in agreement with the observation that flanking an insert with LNA modifications did not completely evade DNA MMR in the oligo-targeting experiments in mESCs (Fig. 3B). We find that LNA modification of a single base mismatch prevents MutS binding and thereby prevents initiation of the MMR pathway.
Fig. 6.
LNA modification of mismatched DNA prevents binding of bacterial MutS protein. (A) Equilibrium binding experiments by FP with purified E. coli MutS using Cy5-labeled 21-bp heteroduplex DNA containing a GT or TT mismatch or an A insert with and without LNA modifications. Measurements are presented as the average of at least three replicates; error bars represent SEM. (B) MutS binding constants (Kdapp) to heteroduplex DNA with and without LNA modification as determined by nonlinear regression fitting of FP data.
Discussion
The use of ssODNs to generate subtle base-pair modifications has been demonstrated in both prokaryotic and eukaryotic cells. Evidence suggests that ssODNs anneal to their complementary target DNA when it is transiently single-stranded during replication. However, MMR represses the efficiency of ssODN-mediated gene modification, thereby severely limiting its applicability (14, 20, 21). Different strategies have been reported to overcome the repressive action of MMR, which include permanent MMR disruption, transient knockdown of MMR activity, and the use of base analogs that attenuate recognition of mismatches by MMR (28–31). Here we present an ssODN design in which strategically placed LNA modifications allow evasion of MMR and hence effective gene modification in MMR-proficient cells.
LNA modifications have been used before to improve the efficacy of ssODN-directed base substitution (11, 42). However, LNAs were placed at the termini of ssODNs with the rationale to improve their intracellular stability. Terminally placed LNAs improved the base-pair substitution efficacy of short (25-nt), but not longer (45-nt), ssODNs, and this effect was solely dependent on the presence of a 5′ end LNA (42). We made a similar observation: The performance of 25-nt, but not 35-nt, ssODNs was improved by a 5′ end, but not 3′ end, LNA. However, we only detected a twofold improvement, whereas Andrieu-Soler et al. (42) achieved 40- to 50-fold increased gene-modification efficiencies. Possibly, in their cell system, ssODNs were more susceptible to 5′ end degradation than in the mESCs we used. The improvement they obtained was certainly not related to an effect on DNA MMR because they made use of the MMR-defective cell line HEK293T (43). Similarly, the efficiency improvement we observed by 5′ acridine modification of LMOs was modest in comparison with observations by others (40), which may also be related to low susceptibility to 5′ end degradation in mESCs.
We demonstrate here that LNAs should be placed at or at and adjacent to the mismatching nucleotides to render ssODNs equally effective in Msh2−/− cells and Msh2+/+ mESCs for the substitution of one, two, and three bases. This finding was extended to λ-red–mediated recombineering in E. coli, where LNA modification of ssODNs strongly enhanced the efficiency of single base pair substitutions at two loci in MMR-proficient cells. Additionally, we demonstrated that LNA modification increased the efficiency of ssODNs for the insertion of one or four bases in MMR-proficient mESCs.
In our experimental setup, we could only test a limited number of all possible single base substitutions. Nevertheless, as a modified sugar group with an additional 2′-C,4′-C-oxymethylene linker differentiates LNA from DNA, we expect LNA-mediated MMR evasion to rely on a general principle that allows MMR evasion for any single base substitution. Consistently, we found a beneficial effect of LNA modification for different compound base substitutions.
In an in vitro binding assay, we found that LNA modification of mismatches severely perturbed mismatch binding by purified MutS protein. In the present MMR model, the weak interaction of MSH proteins with DNA is stabilized when a mismatch is encountered (44, 45). This stabilization is accompanied by bending of the DNA and conformational changes in the MSH dimer that, upon exchange of ADP for ATP, lead to the formation of a sliding clamp (44). The effects of locked nucleotides on backbone flexibility and base stacking may render mismatches in the direct vicinity of LNAs refractory to recognition by MMR proteins. LNA nucleotides are confined to the 3′-endo conformation and induce a shift in sugar puckering toward this conformation of directly neighboring nucleotides, which is most pronounced at the 3′ side (46, 47). Thus, LNAs near a mismatch may prohibit the bending of the DNA that is required for stabilization of the MutS–DNA interaction.
In conclusion, our results demonstrate that incorporation of LNA nucleotides in ssODNs at mismatching bases prevents MMR-mediated suppression of gene modification in mESCs and E. coli. Together with improved oligonucleotide delivery, optimization of the oligonucleotide length, and the addition of a 5′ acridine moiety, LNA modification of ssODNs increased the targeting efficiency in MMR-proficient mESCs from <10−7 to >10−3. In the past, we have used oligonucleotide-directed gene modification to target endogenous genes in mESCs. Previously targeted genes include RB1 (48), FANCF (36), MYCN (28), MSH2 (49, 50), and MSH6 (51). Although successful gene modification in all these cases necessitated transient suppression of MMR, they demonstrate that endogenous genes are accessible to base substitution by single-stranded DNA oligonucleotides. It is most likely that these cases will benefit from LNA modification.
The frequency of oligo targeting remained lower than recently reported frequencies of CRISPR/Cas9-assisted gene editing (2). Nonetheless, because of its high precision, simplicity, and cost-effectiveness, we believe LMO-directed gene modification is an attractive strategy in cases where large numbers of sequence variants need to be evaluated, and their phenotype can be scored by an easily detectable readout. Along this line, we have used oligo targeting to identify pathogenic variants of the key MMR gene MSH2 (55).
Materials and Methods
Oligo Targeting in mESCs.
To assess the efficiency of ssODN-directed gene modification, we used two published, stably integrated selectable mutant neo in wild-type (Msh2+/+) and Msh2−/− mESCs (20). Details for the oligo-targeting protocol are provided in SI Materials and Methods. Briefly, cells were seeded onto six-well plates at a density of 7 × 105 and were exposed on the next day to 1.4 mL of complete medium without serum containing 3 μg of ssODNs and 27 μL of Transfast (Promega) for 75 min at 37 °C. After the addition of 4 mL of 60% BRL-conditioned medium, cells were incubated overnight. Then, cells were counted and reseeded onto 10-cm plates. Finally, successfully modified clones were selected with 750 μg/mL G418 (Geneticin; Life Technologies). In an optimized protocol (Fig. 4), transfection was prepared with 7.5 µL of TransIT-siQUEST (Mirus) and 3 µg of ssODN (unless otherwise indicated).
Recombination Assays in E. coli.
MMR-proficient strain (mutS+) HME6 is W3310 galKTYR145UAG ∆lacU169 Gal+{λcI857∆cro-bioA}, and MMR-deficient strain (∆mutS) HME63 is HME6 mutS<>amp and were provided by Donald L. Court, National Cancer Institute at Frederick, Frederick, MD (13, 14). λ-Red functions were induced at 42 °C for 15 min, after which cells were immediately made electrocompetent as described (52). For GalK correction experiments, we used freshly prepared competent cells, whereas we used competent cells stored at −80 °C in 12.5% (vol/vol) glycerol for modification of rpoB. After electroporation with 5 pmol of ssODN, cells were recovered for either 30 min or overnight at 32 °C in LB for galK and rpoB modification, respectively. Gal+ recombinants were selected on M63 minimal galactose plates with biotin. rpoB D516N mutants were selected on LB agar with 100 μg/mL rifampicin (Sigma-Aldrich). To determine total viable cell count, cells were also plated on LB agar.
DNA Binding of MutS.
A 5′ Cy5-labeled oligonucleotide of 21 residues was annealed to different complementary oligonucleotides to generate either a GT or TT mismatch or an extrahelical A with and without LNA modifications (see Table S1 for oligonucleotide sequences). FP measurements to determine the DNA binding affinities of purified E. coli MutS protein (53) to various mismatches were performed in 50 μL of buffer containing 25 mM Hepes (pH 7.5), 150 mM KCl, 5 mM MgCl2, 0.005% Tween, and 2 or 4 nM fluorescent DNA probe (for, respectively, T:T minus LNA, homoduplex minus LNA, and all other probes). MutS protein was combined with the probe and subsequently serial diluted in black flat-bottom 96-well plates. After 5-min equilibration at room temperature, polarization was measured in a PHERAstar FS machine (BMG Labtech) with a 590/675 (excitation/emission) FP module. GraphPad Prism (GraphPad Software) was used to determine Kdapp values using nonlinear regression fitting with a model that corrects for depletion of the protein (54).
Table S1.
Oligonucleotides are listed per figure
| Oligonucleotides | Sequence 5′–3′ |
| Fig. 1 | |
| xmx | TTCTAGAGCCGCCACCATGGCCTATGCATCGAGCT |
| xMx | TTCTAGAGCCGCCACCA{T}GGCCTATGCATCGAGCT |
| XmX | TTCTAGAGCCGCCACC{A}T{G}GCCTATGCATCGAGCT |
| XMX | TTCTAGAGCCGCCACC{ATG}GCCTATGCATCGAGCT |
| X..xmx..X | {T}TCTAGAGCCGCCACCATGGCCTATGCATCGAGC{T} |
| X..mXm..X | {T}TCTAGAGCCGCCACCA{T}GGCCTATGCATCGAGC{T} |
| Fig. 2 | |
| xmmx | AATTCTAGAGCCGCCATGAAGGCCTATGCATCGAG |
| xMmx | AATTCTAGAGCCGCCA{T}GAAGGCCTATGCATCGAG |
| xmMx | AATTCTAGAGCCGCCAT{G}AAGGCCTATGCATCGAG |
| xMMx | AATTCTAGAGCCGCCA{TG}AAGGCCTATGCATCGAG |
| XmMx | AATTCTAGAGCCGCC{A}T{G}AAGGCCTATGCATCGAG |
| xMmX | AATTCTAGAGCCGCCA{T}G{A}AGGCCTATGCATCGAG |
| XmmX | AATTCTAGAGCCGCC{A}TG{A}AGGCCTATGCATCGAG |
| XMMX | AATTCTAGAGCCGCC{ATGA}AGGCCTATGCATCGAG |
| xMxMx | CTAGAGCCGCCACCA{T}G{T}CCTATGCATCGAGCTTG |
| XMXMX | CTAGAGCCGCCACC{ATGTC}CTATGCATCGAGCTTG |
| Fig. S1 | |
| ammma | AATTCTAGAGCCGCCATGGAGGCCTATGCATCGAGCTT |
| aMMMa | AATTCTAGAGCCGCCA{TGG}AGGCCTATGCATCGAGCTT |
| AMMMA | AATTCTAGAGCCGCC{ATGGA}GGCCTATGCATCGAGCTT |
| gmmmt | CTAGAGCCGCCACCAAGATGTATGCATCGAGCTTG |
| gMMMt | CTAGAGCCGCCACCAAG{ATG}TATGCATCGAGCTTG |
| GMMMT | CTAGAGCCGCCACCAA{GATGT}ATGCATCGAGCTTG |
| cmmmg | GAGCCGCCACCAAGGCCATGGCATCGAGCTTGGATGG |
| cMMMg | GAGCCGCCACCAAGGCC{ATG}GCATCGAGCTTGGATGG |
| CMMMG | GAGCCGCCACCAAGGC{CATGG}CATCGAGCTTGGATGG |
| Fig. 3 | |
| xix | CTAGAGCCGCCACCATGGATATCACCGATGCATCG |
| XiX | CTAGAGCCGCCACCATG{G}A{T}ATCACCGATGCATCG |
| XIX | CTAGAGCCGCCACCATG{GAT}ATCACCGATGCATCG |
| xiiiix | TCTAGAGCCGCCACCATGCTACGTATCACCGATGCATCG |
| XiiiiX | TCTAGAGCCGCCACCAT{G}CTAC{G}TATCACCGATGCATCG |
| XIIIIX | TCTAGAGCCGCCACCAT{GCTACG}TATCACCGATGCATCG |
| Fig. 4A | |
| 17 nt | CGCCACCA{T}GGCCTATG |
| 21 nt | GCCGCCACCA{T}GGCCTATGCA |
| 25 nt | GAGCCGCCACCA{T}GGCCTATGCATC |
| 29 nt | TAGAGCCGCCACCA{T}GGCCTATGCATCGA |
| 35 nt | TTCTAGAGCCGCCACCA{T}GGCCTATGCATCGAGCT |
| 39 nt | AATTCTAGAGCCGCCACCA{T}GGCCTATGCATCGAGCTTG |
| 45 nt | CCCAATTCTAGAGCCGCCACCA{T}GGCCTATGCATCGAGCTTGGAT |
| 55 nt | AGCGACCCAATTCTAGAGCCGCCACCA{T}GGCCTATGCATCGAGCTTGGATGGATT |
| Fig. 4B | |
| 25 nt | GAGCCGCCACCA{T}GGCCTATGCATC |
| 5′LNA | {G}AGCCGCCACCA{T}GGCCTATGCATC |
| 3′LNA | GAGCCGCCACCA{T}GGCCTATGCAT{C} |
| 5′+3′LNA | {G}AGCCGCCACCA{T}GGCCTATGCAT{C} |
| 7x LNA | {G}AGC{C}GCC{A}CCA{T}GGC{C}TAT{G}CAT{C} |
| Fig. 4C | |
| Unmodified | GAGCCGCCACCA{T}GGCCTATGCATC |
| LNA | {G}AGCCGCCACCA{T}GGCCTATGCATC |
| Acr | Acr-GAGCCGCCACCA{T}GGCCTATGCATC |
| Acr+LNA | Acr-{G}AGCCGCCACCA{T}GGCCTATGCATC |
| Fig. 4D | |
| 17 nt | Acr-CGCCACCA{T}GGCCTATG |
| 21 nt | Acr-GCCGCCACCA{T}GGCCTATGCA |
| 25 nt | Acr-GAGCCGCCACCA{T}GGCCTATGCATC |
| 35 nt | Acr-TTCTAGAGCCGCCACCA{T}GGCCTATGCATCGAGCT |
| Fig. 5A | |
| xmx | TTTGTGCGTAATCTCAGACAGCGGGTTGTTCTGGTTCATAAACTGAGACAGCTGGCTGGAACCGAAGAAC |
| xMx | TTTGTGCGTAATCTCAGACAGCGGGTTGTTCTGGT{T}CATAAACTGAGACAGCTGGCTGGAACCGAAGAAC |
| Fig. 5B | |
| xmx | AAGTCGCGGTCGGAACCGTATTGCAGCAGCTTTATCATCTGCCGCTGGACGGCGCACAAATCGCGCTTAA |
| xMx | AAGTCGCGGTCGGAACCGTATTGCAGCAGCTTTA{T}CATCTGCCGCTGGACGGCGCACAAATCGCGCTTAA |
| Fig. 6 | |
| Cy5 FP probe | Cy5-TGCATAGGACTTGGTGGCGGC |
| Homoduplex | GCCGCCACCAAGTCCTATGCA |
| Homoduplex+LNA | GCCGCCACCA{A}GTCCTATGCA |
| TT MM | GCCGCCACCATGTCCTATGCA |
| TT MM+LNA | GCCGCCACCA{T}GTCCTATGCA |
| GT MM | GCCGCCACCAGGTCCTATGCA |
| GT MM+LNA | GCCGCCACCA{G}GTCCTATGCA |
| A insert | GCCGCCACCAAGATCCTATGCA |
| A insert+LNA | GCCGCCACCAA{G}A{T}CCTATGCA |
Modifications are indicated as follows: { }, LNA-modified base; Acr, 9-amino-6-chloro-2-methoxy-acridine; Cy5, Cy5 fluorescent moiety.
Materials.
All oligonucleotides were supplied by Eurogentec; a complete list is provided in Table S1. Those used for mESC experiments were delivered salt-free but otherwise unpurified and dissolved in PBS at a concentration of 1 μg/μL. ssODNs for E. coli experiments were purified by reverse-phase chromatography and dissolved in ddH2O. Oligonucleotides used for FP experiments were reverse-phase HPLC-purified and were diluted and subsequently annealed in 25 mM Hepes (pH 7.5), 150 mM KCl, and 5 mM EDTA. The annealed oligonucleotides were purified by using anion exchange chromatography on a Mini Q PC 3.2/3 Precision Column (GE).
SI Materials and Methods
ESCs were routinely cultured on top of a feeder layer of irradiated mouse embryonic fibroblasts in complete medium (27). For oligo-targeting experiments, cells were seeded onto gelatin-coated six-well plates at a density of 7 × 105 per well and cultured in 60% BRL-conditioned medium (27). The next day, the cells were exposed to 1.4 mL of complete medium without serum containing 3 μg of ssODNs (1 μg/μL) and 27 μL of Transfast (Promega) for 75 min at 37 °C (20). After the addition of 4 mL of 60% BRL-conditioned medium, cells were incubated overnight. Cells were counted, reseeded onto 10-cm gelatin-coated plates at a density of 1–2 × 106 cells per plate, and cultured overnight in 30% BRL-conditioned medium. The next day, selection for successfully modified clones started by adding 750 μg/mL G418 (Geneticin; Life Technologies). The number of surviving colonies was determined 12 d after transfection. Targeting efficiencies were expressed as the ratio of the number of G418-resistant colonies over the number of seeded cells after ssODN transfection. Because targeting efficiencies can vary severalfold between independent experiments, we chose not to provide SDs of targeting efficiencies, but rather to calculate the mean fold difference and corresponding SDs between Msh2−/− and Msh2+/+ cells obtained from independent experiments.
In an optimized protocol (Fig. 4), transfection medium was prepared with 250 μL of complete medium without serum, 7.5 µL of TransIT-siQUEST (Mirus), and 3 µg of ssODN (unless otherwise indicated) and incubated for 15 min at room temperature. Cells on gelatin-coated six-well plates (seeded the previous day at 7 × 105 per well) were refreshed with 3 mL of 60% BRL-conditioned medium, and transfection mix was added dropwise. After 24 h of incubation at 37 °C, the protocol continued as described above.
Detailed protocols for mESC culturing can be found in Aarts et al. (27).
Acknowledgments
We thank Dr. Donald L. Court for sharing HME6 and HME63 strains; and our colleagues Hellen Houlleberghs and Tim Harmsen for useful discussions and comments on the manuscript. This work was supported by Dutch Organization for Research Grant NWO-ALW 822.02.010.
Footnotes
The authors declare no conflict of interest.
This article is a PNAS Direct Submission.
See Commentary on page 3918.
This article contains supporting information online at www.pnas.org/lookup/suppl/doi:10.1073/pnas.1513315113/-/DCSupplemental.
References
- 1.Doudna JA, Charpentier E. The new frontier of genome engineering with CRISPR-Cas9. Science. 2014;346(6213):1258096–1258096. doi: 10.1126/science.1258096. [DOI] [PubMed] [Google Scholar]
- 2.González F, et al. An iCRISPR platform for rapid, multiplexable, and inducible genome editing in human pluripotent stem cells. Cell Stem Cell. 2014;15(2):215–226. doi: 10.1016/j.stem.2014.05.018. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Aarts M, te Riele H. Progress and prospects: Oligonucleotide-directed gene modification in mouse embryonic stem cells: A route to therapeutic application. Gene Ther. 2011;18(3):213–219. doi: 10.1038/gt.2010.161. [DOI] [PubMed] [Google Scholar]
- 4.Li XT, et al. Identification of factors influencing strand bias in oligonucleotide-mediated recombination in Escherichia coli. Nucleic Acids Res. 2003;31(22):6674–6687. doi: 10.1093/nar/gkg844. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Brachman EE, Kmiec EB. Gene repair in mammalian cells is stimulated by the elongation of S phase and transient stalling of replication forks. DNA Repair (Amst) 2005;4(4):445–457. doi: 10.1016/j.dnarep.2004.11.007. [DOI] [PubMed] [Google Scholar]
- 6.Olsen PA, Randol M, Krauss S. Implications of cell cycle progression on functional sequence correction by short single-stranded DNA oligonucleotides. Gene Ther. 2005;12(6):546–551. doi: 10.1038/sj.gt.3302454. [DOI] [PubMed] [Google Scholar]
- 7.Aarts M, te Riele H. Subtle gene modification in mouse ES cells: Evidence for incorporation of unmodified oligonucleotides without induction of DNA damage. Nucleic Acids Res. 2010;38(20):6956–6967. doi: 10.1093/nar/gkq589. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Huen MSY, et al. The involvement of replication in single stranded oligonucleotide-mediated gene repair. Nucleic Acids Res. 2006;34(21):6183–6194. doi: 10.1093/nar/gkl852. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Radecke S, Radecke F, Peter I, Schwarz K. Physical incorporation of a single-stranded oligodeoxynucleotide during targeted repair of a human chromosomal locus. J Gene Med. 2006;8(2):217–228. doi: 10.1002/jgm.828. [DOI] [PubMed] [Google Scholar]
- 10.Wu Q, Christensen LA, Legerski RJ, Vasquez KM. Mismatch repair participates in error-free processing of DNA interstrand crosslinks in human cells. EMBO Rep. 2005;6(6):551–557. doi: 10.1038/sj.embor.7400418. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Aarts M, te Riele H. Parameters of oligonucleotide-mediated gene modification in mouse ES cells. J Cell Mol Med. 2010;14(6B):1657–1667. doi: 10.1111/j.1582-4934.2009.00847.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Yu D, et al. An efficient recombination system for chromosome engineering in Escherichia coli. Proc Natl Acad Sci USA. 2000;97(11):5978–5983. doi: 10.1073/pnas.100127597. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Ellis HM, Yu D, DiTizio T, Court DL. High efficiency mutagenesis, repair, and engineering of chromosomal DNA using single-stranded oligonucleotides. Proc Natl Acad Sci USA. 2001;98(12):6742–6746. doi: 10.1073/pnas.121164898. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Costantino N, Court DL. Enhanced levels of lambda Red-mediated recombinants in mismatch repair mutants. Proc Natl Acad Sci USA. 2003;100(26):15748–15753. doi: 10.1073/pnas.2434959100. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Carter DM, Radding CM. The role of exonuclease and beta protein of phage lambda in genetic recombination. II. Substrate specificity and the mode of action of lambda exonuclease. J Biol Chem. 1971;246(8):2502–2512. [PubMed] [Google Scholar]
- 16.Kmiec E, Holloman WK. Beta protein of bacteriophage lambda promotes renaturation of DNA. J Biol Chem. 1981;256(24):12636–12639. [PubMed] [Google Scholar]
- 17.Muniyappa K, Radding CM. The homologous recombination system of phage lambda. Pairing activities of beta protein. J Biol Chem. 1986;261(16):7472–7478. [PubMed] [Google Scholar]
- 18.Mythili E, Kumar KA, Muniyappa K. Characterization of the DNA-binding domain of beta protein, a component of phage lambda red-pathway, by UV catalyzed cross-linking. Gene. 1996;182(1-2):81–87. doi: 10.1016/s0378-1119(96)00518-5. [DOI] [PubMed] [Google Scholar]
- 19.Karakousis G, et al. The beta protein of phage lambda binds preferentially to an intermediate in DNA renaturation. J Mol Biol. 1998;276(4):721–731. doi: 10.1006/jmbi.1997.1573. [DOI] [PubMed] [Google Scholar]
- 20.Dekker M, Brouwers C, te Riele H. Targeted gene modification in mismatch-repair-deficient embryonic stem cells by single-stranded DNA oligonucleotides. Nucleic Acids Res. 2003;31(6):e27. doi: 10.1093/nar/gng027. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Kow YW, Bao G, Reeves JW, Jinks-Robertson S, Crouse GF. Oligonucleotide transformation of yeast reveals mismatch repair complexes to be differentially active on DNA replication strands. Proc Natl Acad Sci USA. 2007;104(27):11352–11357. doi: 10.1073/pnas.0704695104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.de Wind N, Dekker M, Berns A, Radman M, te Riele H. Inactivation of the mouse Msh2 gene results in mismatch repair deficiency, methylation tolerance, hyperrecombination, and predisposition to cancer. Cell. 1995;82(2):321–330. doi: 10.1016/0092-8674(95)90319-4. [DOI] [PubMed] [Google Scholar]
- 23.Reitmair AH, et al. MSH2 deficient mice are viable and susceptible to lymphoid tumours. Nat Genet. 1995;11(1):64–70. doi: 10.1038/ng0995-64. [DOI] [PubMed] [Google Scholar]
- 24.Baker SM, et al. Involvement of mouse Mlh1 in DNA mismatch repair and meiotic crossing over. Nat Genet. 1996;13(3):336–342. doi: 10.1038/ng0796-336. [DOI] [PubMed] [Google Scholar]
- 25.Edelmann W, et al. Meiotic pachytene arrest in MLH1-deficient mice. Cell. 1996;85(7):1125–1134. doi: 10.1016/s0092-8674(00)81312-4. [DOI] [PubMed] [Google Scholar]
- 26.Edelmann W, et al. Mutation in the mismatch repair gene Msh6 causes cancer susceptibility. Cell. 1997;91(4):467–477. doi: 10.1016/s0092-8674(00)80433-x. [DOI] [PubMed] [Google Scholar]
- 27.Aarts M, et al. Gene modification in embryonic stem cells by single-stranded DNA oligonucleotides. Methods Mol Biol. 2009;530:79–99. doi: 10.1007/978-1-59745-471-1_5. [DOI] [PubMed] [Google Scholar]
- 28.Dekker M, et al. Transient suppression of MLH1 allows effective single-nucleotide substitution by single-stranded DNA oligonucleotides. Mutat Res. 2011;715(1-2):52–60. doi: 10.1016/j.mrfmmm.2011.07.008. [DOI] [PubMed] [Google Scholar]
- 29.Nyerges Á, et al. Conditional DNA repair mutants enable highly precise genome engineering. Nucleic Acids Res. 2014;42(8):e62. doi: 10.1093/nar/gku105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Wang HH, Xu G, Vonner AJ, Church G. Modified bases enable high-efficiency oligonucleotide-mediated allelic replacement via mismatch repair evasion. Nucleic Acids Res. 2011;39(16):7336–7347. doi: 10.1093/nar/gkr183. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Rios X, et al. Stable gene targeting in human cells using single-strand oligonucleotides with modified bases. PLoS One. 2012;7(5):e36697. doi: 10.1371/journal.pone.0036697. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Obika S, et al. Synthesis of 2′-O,4′-C-methyleneuridine and -cytidine. Novel bicyclic nucleosides having a fixed C3, -endo sugar puckering. Tetrahedron Lett. 1997;38(50):8735–8738. [Google Scholar]
- 33.Singh SK, Koshkin AA, Wengel J, Nielsen P. LNA (locked nucleic acids): Synthesis and high-affinity nucleic acid recognition. Chem Commun (Camb) 1998;(4):455–456. [Google Scholar]
- 34.Obika S, et al. Stability and structural features of the duplexes containing nucleoside analogues with a fixed N-type conformation, 2′-O,4′-C-methyleneribonucleosides. Tetrahedron Lett. 1998;39(30):5401–5404. [Google Scholar]
- 35.Koshkin AA, et al. LNA (locked nucleic acids): Synthesis of the adenine, cytosine, guanine, 5-methylcytosine, thymine and uracil bicyclonucleoside monomers, oligomerisation, and unprecedented nucleic acid recognition. Tetrahedron. 1998;54(14):3607–3630. [Google Scholar]
- 36.Dekker M, et al. Effective oligonucleotide-mediated gene disruption in ES cells lacking the mismatch repair protein MSH3. Gene Ther. 2006;13(8):686–694. doi: 10.1038/sj.gt.3302689. [DOI] [PubMed] [Google Scholar]
- 37.Drummond JT, Li GM, Longley MJ, Modrich P. Isolation of an hMSH2-p160 heterodimer that restores DNA mismatch repair to tumor cells. Science. 1995;268(5219):1909–1912. doi: 10.1126/science.7604264. [DOI] [PubMed] [Google Scholar]
- 38.Palombo F, et al. hMutSbeta, a heterodimer of hMSH2 and hMSH3, binds to insertion/deletion loops in DNA. Curr Biol. 1996;6(9):1181–1184. doi: 10.1016/s0960-9822(02)70685-4. [DOI] [PubMed] [Google Scholar]
- 39.Brunet E, et al. Intercalator conjugates of pyrimidine locked nucleic acid-modified triplex-forming oligonucleotides: Improving DNA binding properties and reaching cellular activities. Nucleic Acids Res. 2005;33(13):4223–4234. doi: 10.1093/nar/gki726. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.de Piédoue G, et al. Targeted gene correction with 5′ acridine-oligonucleotide conjugates. Oligonucleotides. 2007;17(2):258–263. doi: 10.1089/oli.2007.0074. [DOI] [PubMed] [Google Scholar]
- 41.Campbell EA, et al. Structural mechanism for rifampicin inhibition of bacterial RNA polymerase. Cell. 2001;104(6):901–912. doi: 10.1016/s0092-8674(01)00286-0. [DOI] [PubMed] [Google Scholar]
- 42.Andrieu-Soler C, et al. Stable transmission of targeted gene modification using single-stranded oligonucleotides with flanking LNAs. Nucleic Acids Res. 2005;33(12):3733–3742. doi: 10.1093/nar/gki686. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Trojan J, et al. Functional analysis of hMLH1 variants and HNPCC-related mutations using a human expression system. Gastroenterology. 2002;122(1):211–219. doi: 10.1053/gast.2002.30296. [DOI] [PubMed] [Google Scholar]
- 44.Cho WK, et al. ATP alters the diffusion mechanics of MutS on mismatched DNA. Structure. 2012;20(7):1264–1274. doi: 10.1016/j.str.2012.04.017. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Lee JB, et al. Single-molecule views of MutS on mismatched DNA. DNA Repair (Amst) 2014;20:82–93. doi: 10.1016/j.dnarep.2014.02.014. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Petersen M, et al. The conformations of locked nucleic acids (LNA) J Mol Recognit. 2000;13(1):44–53. doi: 10.1002/(SICI)1099-1352(200001/02)13:1<44::AID-JMR486>3.0.CO;2-6. [DOI] [PubMed] [Google Scholar]
- 47.Ivanova A, Rösch N. The structure of LNA:DNA hybrids from molecular dynamics simulations: The effect of locked nucleotides. J Phys Chem A. 2007;111(38):9307–9319. doi: 10.1021/jp073198j. [DOI] [PubMed] [Google Scholar]
- 48.Aarts M, Dekker M, de Vries S, van der Wal A, te Riele H. Generation of a mouse mutant by oligonucleotide-mediated gene modification in ES cells. Nucleic Acids Res. 2006;34(21):e147. doi: 10.1093/nar/gkl896. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Wielders EA, Dekker RJ, Holt I, Morris GE, te Riele H. Characterization of MSH2 variants by endogenous gene modification in mouse embryonic stem cells. Hum Mutat. 2011;32(4):389–396. doi: 10.1002/humu.21448. [DOI] [PubMed] [Google Scholar]
- 50.Wielders EA, et al. Functional analysis of MSH2 unclassified variants found in suspected Lynch syndrome patients reveals pathogenicity due to attenuated mismatch repair. J Med Genet. 2014;51(4):245–253. doi: 10.1136/jmedgenet-2013-101987. [DOI] [PubMed] [Google Scholar]
- 51.Wielders EA, Houlleberghs H, Isik G, te Riele H. Functional analysis in mouse embryonic stem cells reveals wild-type activity for three MSH6 variants found in suspected Lynch syndrome patients. PLoS One. 2013;8(9):e74766. doi: 10.1371/journal.pone.0074766. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Sawitzke JA, et al. Recombineering: Highly efficient in vivo genetic engineering using single-strand oligos. Methods Enzymol. 2013;533:157–177. doi: 10.1016/B978-0-12-420067-8.00010-6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Lamers MH, et al. The crystal structure of DNA mismatch repair protein MutS binding to a G x T mismatch. Nature. 2000;407(6805):711–717. doi: 10.1038/35037523. [DOI] [PubMed] [Google Scholar]
- 54.Lundblad JR, Laurance M, Goodman RH. Fluorescence polarization analysis of protein-DNA and protein-protein interactions. Mol Endocrinol. 1996;10(6):607–612. doi: 10.1210/mend.10.6.8776720. [DOI] [PubMed] [Google Scholar]
- 55.Houlleberghs H, et al. An oligonucleotide-directed mutagenesis screen to identify pathogenic Lynch syndrome associated MSH2 DNA mismatch repair gene variants. Proc Natl Acad Sci USA. 2016;113:4128–4133. doi: 10.1073/pnas.1520813113. [DOI] [PMC free article] [PubMed] [Google Scholar]