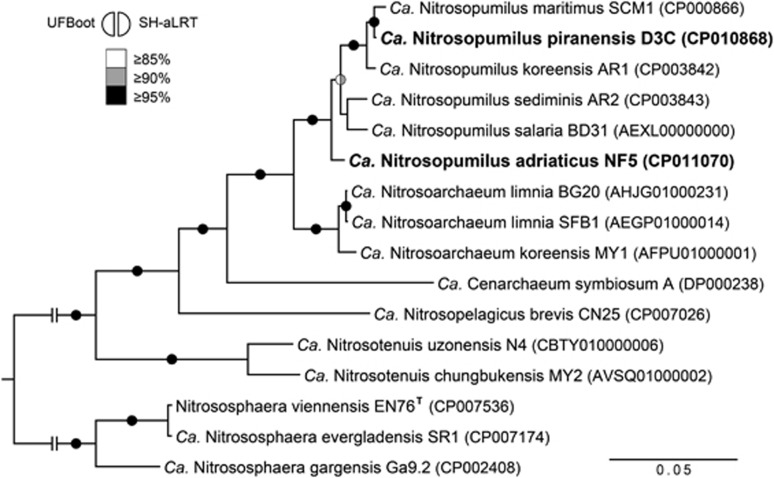
Figure 1.
Phylogenetic tree of concatenated 16S–23S rRNA genes from cultivated Thaumarchaeota with sequenced genomes (16 organisms) showing the affiliation of strains D3C and NF5. Full-length gene sequences were aligned with MAFFT (L-INS-I method) based on Archaea-specific structurally accurate seed alignments (1474 and 3026 aligned positions for 16S and 23S rRNA genes, respectively), and unreliable positions were filtered from the resulting alignments with TCS before concatenation (4443 aligned positions in the concatenated alignment). The tree was calculated by maximum likelihood with IQ-Tree based on the GTR+I+Γ4 model, with ultrafast bootstrap (UFBoot) and SH-aLRT support values inferred from 1000 replicates each (see Supplementary materials and methods for details). Support values ⩾85% are represented on the respective branches by semi-circles color-coded as indicated on the figure.