Figure 2.
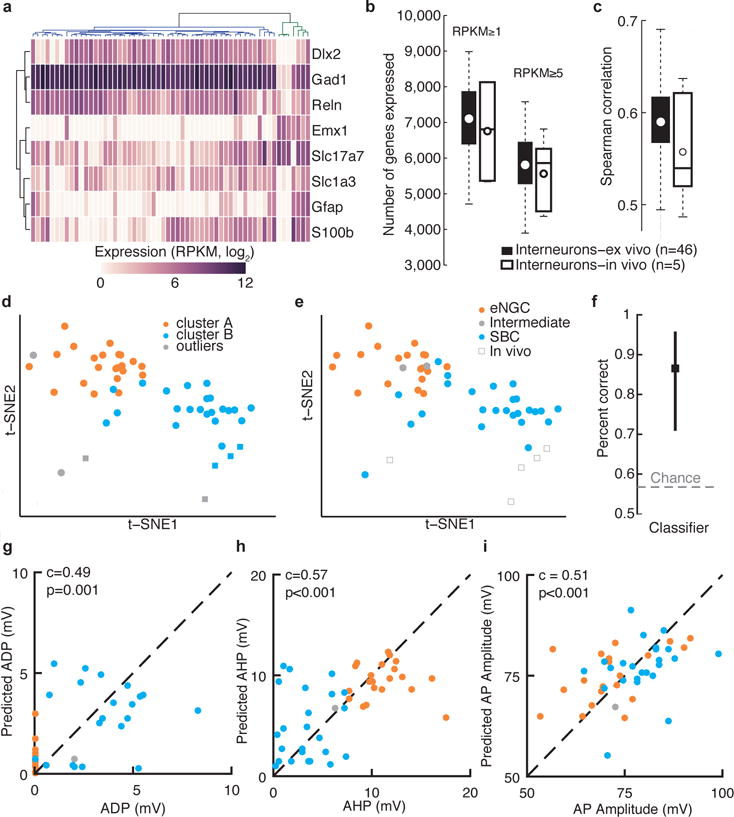
Single-neuron transcriptome profiles predict cell type and electrophysiological properties. (a) Clustering analysis separates interneurons (blue dendrogram subtree) from other neuronal classes (green dendrogram subtree; includes four pyramidal neurons, and one astrocyte) based on marker gene expression. Two L1 interneurons clustered with non-interneuron cell types, indicating possible contamination of these samples, and so these two cells were excluded from our analysis of interneuron subtypes. (b) Number of genes detected per neuron using two different expression thresholds for interneurons patch-clamp recorded ex vivo and in vivo. (c) Pairwise Spearman correlation across all detected genes for ex vivo and in vivo patch-clamp recorded interneurons. (d) Two-dimensional t-SNE representation of gene expression for all L1 interneurons. Cells are colored according to affinity propagation-based clustering in gene-space spanned by the 3,000 most variable genes, prior to dimensionality reduction. (e) The same two-dimensional map as in d, but with cells color-coded according to expert classification of cell type based on electrophysiological properties. Performance of GLMs using single-neuron gene expression to predict cell type (f), ADP (g), AHP (h), or action potential (AP) amplitude (i).
