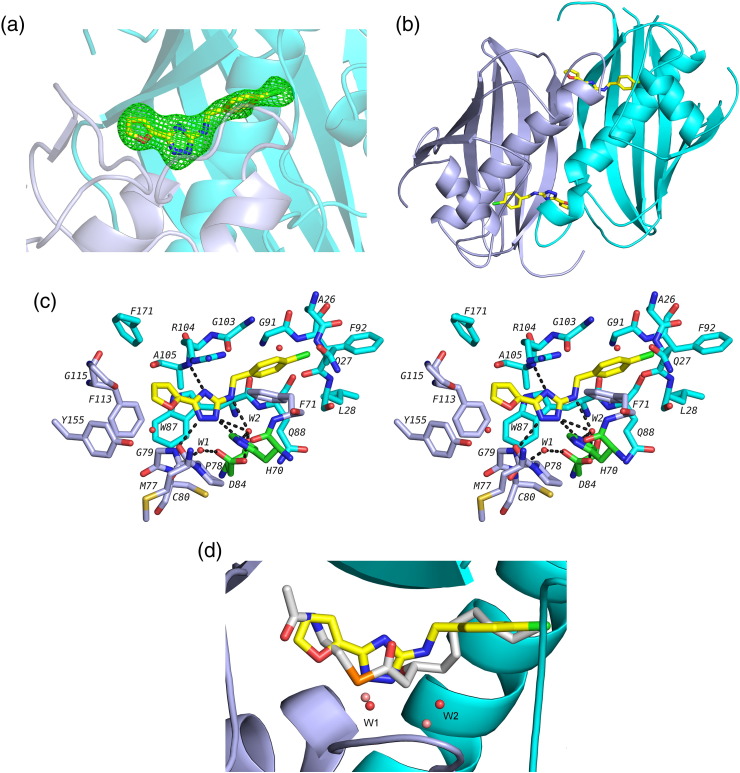
Fig. 6.
Crystal structure of PaFabA complex with N42FTA.
(a) Fo − Fc electron density omit map at 3σ around the N42FTA. The compound is shown as sticks with carbon atoms coloured yellow, chlorine coloured green, nitrogen coloured blue, and oxygen coloured red. The A subunit of the protein is coloured cyan and the B subunit is coloured pale blue.
(b) Two molecules of N42FTA bind to the PaFabA dimer, and each N42FTA molecule binds at the protein dimer interface. The colour scheme is as (a).
(c) Stereo view of N42FTA binding site. Residues within 4.0 Å of the compound are displayed and hydrogen bonds are shown as black broken lines. The colour scheme is as (a), with the carbons from the A subunit coloured cyan and those from the B subunit coloured pale blue. The conserved water, W1, labelled and the additional water (found in all but two subunits) is marked W2. A ligplot diagramme for the interaction is given in Fig. S4. The two catalytic residues (H70 and D84) have been highlighted in green.
(d) Superposition of the N42FTA PaFabA complex with the previously reported 3-hydroxydecanoyl-NAC PaFabA complex [24] reveals that the compound matches the volume of the substrate mimic very well. The colour scheme is as (a) for the N42FTA PaFabA complex but the carbons of 3-hydroxydecanoyl-NAC are coloured grey, the S atom is in orange, and the two water molecules are in pink.