Abstract
The aim of this study was to investigate human invasive isolates of enterococci, obtained through prospective surveillance in Poland. The consecutive enterococcal isolates were collected in 30 hospitals between May 2010 and June 2011, and studied by species identification, antimicrobial susceptibility testing and, for Enterococcus faecium by detection of markers specific for the hospital meroclone, multilocus VNTR analysis (MLVA) and multilocus sequence typing (MLST). Additionally, the genomic difference regions (GDRs) characteristic for lineage 78 were searched by PCR. Among 259 isolates, a nearly equal number of Enterococcus faecalis (n = 140; 54.1 %) and E. faecium (n = 112; 43.2 %) was found. The observed 14-day mortality rate of infected patients reached 18.1 %. All isolates were susceptible to linezolid and daptomycin. High-level aminoglycoside resistance occurred in over 50 % of isolates. Vancomycin resistance mediated by vanA or vanB was detected in 7.1 % of E. faecium; 71.4 % of isolates were multidrug resistant. E. faecium isolates ubiquitously carried molecular markers of hospital-associated meroclone (IS16, espEfm, intA of ICEEfm1) and multilocus sequence typing showed the domination of representatives of lineages 78 and 17/18 (52.7 % and 46.4 %, respectively). Isolates of lineage 78 were significantly enriched in all the GDRs studied. The recent spread of E. faecium from this lineage contributed to the observed increase of E. faecium in enterococcal invasive infections in hospitals in Poland.
Introduction
Bacteria belonging to the genus Enterococcus are a part of the normal, harmless faecal flora of humans and animals. However, they may also act as opportunistic pathogens, and today are recognized as important causative agents of both invasive and non-invasive nosocomial infections, affecting immunocompromised, severely ill patients [1]. According to the European Centre for Disease Prevention and Control (ECDC) enterococci are presently third, after Escherichia coli and Staphylococcus aureus, most frequently isolated bacteria from the healthcare-associated infections (HAIs) in Europe [2]. Currently observed acquisition of resistance to antimicrobials used in the therapy of enterococcal infections is a cause of growing concern [3]. Enterococcus faecalis and Enterococcus faecium represent two species of the biggest clinical importance, while infections caused by other enterococci are rare [4]. Multilocus sequence typing (MLST) and other typing methods showed the presence of distinct clones associated with hospital infections/outbreaks, named high-risk enterococcal clonal complexes (HiRECCs), within populations of both E. faecalis and E. faecium [5, 6]. Genomes of isolates belonging to HiRECCs are enriched in mobile genetic elements (MGEs), often associated with genes encoding potential virulence factors and resistance determinants [3, 7]. For E. faecalis, two HIRECCs, CC6 (also known as CC2) and CC87, preferentially group hospital clinical isolates [8]. Hospital subpopulation of E. faecium, initially described as CC17, was subsequently divided into three lineages, 17, 18, and 78, named after the major sequence type (ST) in each group [9, 10]. Analysis of the MLST data using so-called Bayesian analysis of population structure (BAPS) delimited two groups in the hospital meroclone, 2–1, and 3–3, corresponding to lineages 78 and 17/18 respectively [10]. Acquisition of ampicillin and ciprofloxacin resistance, and the presence of the transferable pathogenicity island (ICEEfm1), harbouring the espEfm gene, are considered important adaptive features of hospital meroclone of E. faecium [11]. Genomic analyses indicated the presence of seven additional regions in the genome of ST203 strain from lineage 78 in comparison to lineage 17/18 [12].
The epidemiology of invasive enterococcal infections in Polish hospitals is monitored by a systematic passive surveillance performed by the National Reference Centre for Susceptibility Testing, located at the National Medicines Institute in Warsaw. However, considering the importance of enterococcal infections, we decided to undertake a study based on enhanced surveillance in selected hospitals during a pre-defined period of time. Objectives of this study were to collect enterococcal isolates from normally sterile body sites together with the relevant clinical data, and to evaluate species distribution and antibiotic susceptibility of these isolates. Additionally, we aimed at more profound phenotypic and molecular characterization of E. faecium isolates, as the clinical importance of this enterococcal species is recently increasing in several countries [13], to search for a possible reason of this phenomenon.
Materials and methods
Bacterial isolates and phenotypic testing
Consecutive invasive isolates of Enterococcus spp. (a single strain per patient from normally sterile body fluids, such as blood, pleural fluid, cerebrospinal fluid, and peritoneal fluid) were collected in 30 collaborating hospitals, located in 26 cities in all regions of Poland, between May 2010 and June 2011. All patient data were collected anonymously in the questionnaire formulated for the purpose of the study. A nosocomial infection was defined as infection which was caused by a strain isolated after 48 hours from admission to the hospital or earlier when patient was transferred from another hospital. Enterococcal isolates were re-identified in the central laboratory by conventional methods, and using the mass spectrometer MALDI Biotyper (Brücker, Bremen, Germany) and Vitek MS (bioMérieux, Marcy l’Etoile, France) for selected isolates. For Enterococcus hirae, Enterococcus durans, and Enterococcus avium identification was conducted additionally with VITEK 2 Compact (bioMérieux). Antimicrobial susceptibility testing for ampicillin, penicillin, imipenem, vancomycin, teicoplanin, gentamicin, streptomycin, linezolid, tigecycline, tetracycline, chloramphenicol, rifampin, ciprofloxacin, quinupristin–dalfopristin and trimethoprim–sulfamethoxazole was performed using the broth microdilution method according to the Clinical and Laboratory Standard Institute guidelines (CLSI), and for daptomycin the Etest method (bioMérieux). The reference strain E. faecalis ATCC 29212 was used for quality control. Obtained results were interpreted using available 2015 breakpoints of The European Committee on Antimicrobial Susceptibility Testing (EUCAST) (http://www.eucast.org/) and the CLSI breakpoints for antimicrobials for which the EUCAST breakpoints were not available. Isolates were considered multidrug-resistant (MDR) when they showed resistance to three or more classes of antimicrobials tested [14]. Biofilm formation was evaluated by the quantitative adherence assay in Trypticase Soy Broth with 0.25 % glucose [15], and isolates were classified as biofilm non-producers, weak and strong producers, based on the results of staining with crystal violet [16].
DNA isolation, gene detection, molecular typing and data analysis
Total DNA was isolated using the Genomic DNA Prep Plus kit according to the manufacturer’s instructions (A&A Biotechnology, Gdynia, Poland). Detection of espEfm and IS16 in E. faecium was performed by PCR as previously described [17, 18]. The intAICEEfm1 gene, encoding the integrase of ICEEfm1 was detected by PCR with primers designed in this study: intA_up2 (5′-AATTGATTCGATAGTTTAGGTA-3') and intA_dn2 (5′-AATCACTTGCTTACTCTTCAT -3′). E. faecium isolates positive for IS16, espEfm and intA from our laboratory collection [19] served as positive controls. Vancomycin nonsusceptibility determinants vanA, vanB, and vanC were detected as previously described [20, 21], with E. faecium BM4147, E. faecalis V583, and Enterococcus gallinarum BM4147 used as respective positive controls. E. faecium isolates were analysed by multiple-locus variable-number tandem repeat (VNTR) analysis (MLVA) according to Top et al. [22] and MLST [5], using the MLST database (http://pubmlst.org/efaecium/) to determine allele numbers and STs (21st April 2015, date accessed). New alleles and allelic profiles were submitted to the above database. MLST data were analyzed with the comparative eBURST analysis against the whole E. faecium database (http://eburst.mlst.net/; 21st April 2015, date accessed). Genomic difference regions (named herein GDRs), differentiating lineages 78 and 17/18 [12], were detected by PCR using primers specific for genes located in these GDRs, based on available genomic sequences (GenBank Accession number NC_021994; Table 1). Differences in distributions were assessed using the chi-squared test with p value ≤ 0.05 considered significant. Antibiotic susceptibility data analysis with the 95 % confidence intervals for the calculation of resistance ratio was done using the WHONET (http://www.whonet.org).
Table 1.
Primers targeting the genomic difference regions
| Primer | Sequence 5′- > 3′ | Gene in AUS0085 (ST203) | Gene product | PCR product size |
|---|---|---|---|---|
| Efm_R1_up | AATCGATGACGTGGAAGAAGG | Ef_aus00245 | cadmium_translocating P-type ATPase | 411 bp |
| Efm_R1_dn | GACTAAAGCGCCAGGACAAC | |||
| Efm_R2_up | ATGTTGCCCAAAAGACGAACC | Ef_aus 01495 | riboflavin biosynthesis protein RibD | 153 bp |
| Efm_R2_dn | GGAACGGCTAAAACAAGAAGC | |||
| Efm_R3_up | GCGTGATTTCGGTAATTGGTG | Ef_aus 02036 | putative phosphosugar isomerase/binding protein | 316 bp |
| Efm_R3_dn | ATGGGAATAGACCAGGAGCA | |||
| Efm_R5_up | CGTGCGTTCCTTTTTCTACC | Ef_aus 02504 | bacteriocin-like protein EntT | 368 bp |
| Efm_R5_dn | GGTTTAGATAGCCCACCAAG | |||
| Efm_R6_up | CCCATGAATCCTGTTGGTTC | Ef_aus 02768 | PTS system, lactose/cellobiose-specific IIC component | 182 bp |
| Efm_R6_dn | GCAAAAGTAGCAGGAAGGAC | |||
| Efm_R7_up | TCAGCAAATGATGGCGATACG | Ef_aus 02778 | glycosyl hydrolase family 38 protein | 374 bp |
| Efm_R7_dn | ACCAATTCGGAGGAATGACATC |
Results
Enterococcal species
Altogether, 259 invasive enterococcal isolates were obtained during the collection period. For 247 (95.4 %) of them, the identification by MALDI Biotyper was consistent with the results obtained with conventional methods and Vitek 2 Compact. For the remaining 12 isolates, additional identification with Vitek MS was used, and its results were in the agreement with the results from MALDI Biotyper in three cases (one isolate of E. faecalis, E. faecium and E. durans each), while in nine cases identification with the Vitek MS confirmed the results of conventional methods (for three E. faecalis, five E. faecium and one E. casseliflavus). Thus, collected isolates comprised 140 strains of E. faecalis (54.1 %), 112 of E. faecium (43.2 %) and seven isolates (2.7 %) of other Enterococcus spp.: three E. gallinarum, two E. durans, and single representatives of E. avium and E. casseliflavus.
Hospitals involved in the study and patients’ data
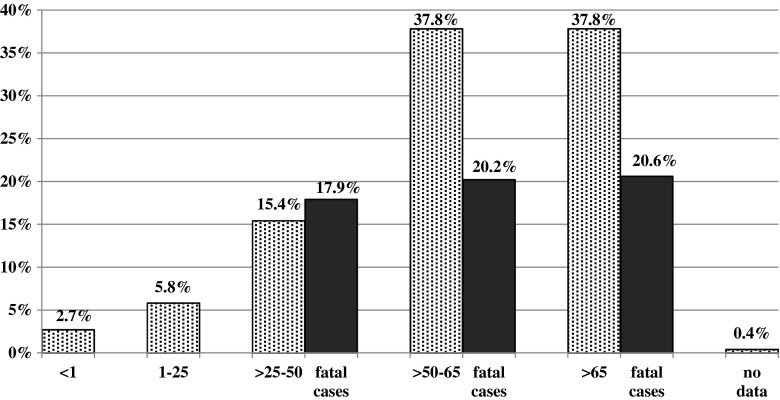
Isolates were obtained from 30 collaborating hospitals (8.6 isolates per hospital). Hospitals involved in the study included secondary and tertiary hospitals, mainly of regional coverage (17 provincial hospitals), but also district hospitals (n = 5), specialist (n = 4) and university (n = 4) hospitals. The numbers of isolates collected from each type of ward are presented in Table 2. For five strains, wards of hospitalization were not given. A significant proportion of enterococcal invasive infections were nosocomial infections, i.e., 65.7 % and 78.6 % of E. faecalis and E. faecium infections respectively. One hundred and forty-five patients (56.0 %) with enterococcal invasive infections were males and 109 (42.1 %) were females; the gender of five patients (1.9 %) was not reported. The age of patients’ ranged from a newborn to 89 years, and the vast majority of isolates (196, i.e., 75.6 %) was obtained from patients aged above 50 years (Fig. 1). The most frequently reported types of infection were bloodstream infections (Table 2), including bacteraemia, septicaemia, and endocarditis (82.9 % of infections caused by E. faecalis and 75.0 % infections caused by E. faecium) followed by abdominal infections (mainly peritonitis), meningitis, and chest and pelvic infections (mainly abscess). Most of the bacteraemia/septicaemia cases had a known focus (E. faecalis 59.3 %, E. faecium 53.6 %). The ratio of bloodstream infections to abdominal infections for E. faecalis was 6.4:1 and for E. faecium 3.8:1, and the observed differences in the ratio of bloodstream infections was statistically significant (p = 0.011).
Table 2.
Ward type, type of infection and outcome reported for patients with invasive enterococcal infections
| E. faecalis (140) | E. faecium (112) | Other species (7) | All | |
|---|---|---|---|---|
| Number of isolates | 140 (54.1 %) | 112 (43.2 %) | 7 | 259 |
| Type of ward | ||||
| Surgery | 32 (22.9 %) | 31 (27.7 %) | 3 | 66 (25.5 %) |
| Haematology/oncology | 20 (14.3 %) | 32 (28.6 %) | 2 | 54 (20.8 %) |
| ICU | 29 (20.7 %) | 27 (24.1 %) | 1 | 57 (22.0 %) |
| Internal medicine | 29 (20.7 %) | 11 (9.8 %) | 0 | 40 (15.4 %) |
| Other neurology (n = 9), dialysis centres (n = 8), neonatal (n = 5), gynaecology (n = 4), urology (n = 5), infectious disease (n = 3), geriatric (n = 2), palliative medicine (n = 1) | 30 (21.4 %) | 11 (9.8 %) | 1 | 42 (16.2 %) |
| Type of infection | ||||
| Bacteraemia, septicaemia, including: | 109 (77.8 %) | 82 (73.2 %) | 4 | 195 (75.3 %) |
| Bacteraemia | 76 (54.3 %) | 63 (56.2 %) | 3 | 142 (54.8 %) |
| Septicaemia | 33 (23.5 %) | 19 (16.9 %) | 1 | 53 (20.5 %) |
| - source abdominal infection | 16 (11.4 %) | 11 (9.8 %) | 2 | 29 (11.2 %) |
| - source urinary tract infection | 10 (7.1 %) | 1 (0.9 %) | 0 | 11 (4.2 %) |
| - other known source | 26 (18.6 %) | 18 (16.1 %) | 0 | 44 (17.0 %) |
| - unknown source | 57 (40.7 %) | 52 (46.4 %) | 0 | 109 (42.1 %) |
| Endocarditis | 7 (5.1 %) | 2 (1.8 %) | 0 | 9 (3.5 %) |
| Abdominal infections | 18 (12.9 %) | 22 (19.6 %) | 3 | 43 (16.6 %) |
| Pleural infections | 2 (1.4 %) | 2 (1.8 %) | 0 | 4 (1.5 %) |
| Meningitis | 2 (1.4 %) | 3 (2.7 %) | 0 | 5 (1.9 %) |
| Pelvic infections | 2 (1.4 %) | 1 (0.9 %) | 0 | 3 (1.2 %) |
| Outcomea | ||||
| All reported | 130 | 107 | 6 | 243 |
| Fatal cases | 20 (15.4 %) | 23 (21.5 %) | 1b | 44 (18.1 %) |
| Cured | 33 (25.4 %) | 25 (23.4 %) | 2 | 60 (24.7 %) |
| Under treatment | 77 (59.2 %) | 59 (55.1 %) | 3 | 139 (57.2 %) |
a% of all cases with the known outcome; b E. gallinarum
In the column for each species the % values were calculated separately taking the number of the isolates belonging the each species as 100 %
Fig. 1.
The age distribution of patients and percentage of fatal cases in each age group
The data concerning risk factors were available for 165 (63.7 %) of patients, and indicated special importance of previous hospitalization during the previous 6 months (n = 124; 75.2 % cases), surgery (n = 42; 25.4 %) and an ICU stay (n = 35; 21.2 %). Only one patient was reported as a nursing home resident. The reported co-morbidities were: malignancy (n = 21; 13.9 %), chronic renal disease (n = 7; 4.6 %), diabetes (n = 4; 2.6 %), and injury (n = 5; 3.3 %). The outcome of treatment of patients with enterococcal infections was reported for 243 cases (93.8 %). Due to the persistence of infection, 2 weeks after strain isolation more than half of the patients (53.7 %) were still receiving antimicrobial treatment, while only 23.2 % of patients were successfully cured. The observed overall 14-day mortality rate reached 18.1 % (44 patients; Table 2). Fatal cases were reported in patients aged above 25 years, and mortality showed an increase with age (Fig. 1). The highest mortality ratio was reported for meningitis (one of three) and septicaemia (30.2 %, 16 of 53). A higher mortality in E. faecium infections (23 cases, 21.5 %) than these caused by E. faecalis (20 cases, 15.4 %) was observed, however, without statistical significance (p = 0.22). A single case of fatal septicaemia in an oncology patient infected with E. gallinarum was reported (Table 2). In two cases, VREm infections proved fatal (28.6 % mortality, i.e., two out of seven cases with the reported outcome), while for vancomycin-susceptible strains this value was 21.0 % (21 of 100 with the reported outcome).
Antimicrobial susceptibility of isolates and biofilm production by E. faecium
All isolates were susceptible to linezolid and daptomycin (Table 3), and all E. faecalis isolates were also susceptible to ampicillin, vancomycin and teicoplanin. Over 50 % of isolates of both species showed high-level resistance to aminoglycosides. Among E. faecium isolates, very high ratios of resistance to ampicillin, ciprofloxacin, and rifampin were observed, and vancomycin resistance was detected in eight isolates (VREfm); five of them showed VanA phenotype and the presence of vanA. The three remaining VREfm carried the vanB gene. Altogether, 71.4 % of isolates were classified as multidrug resistant (MDR) [14]. This phenotype was specific for nearly all isolates of E. faecium (109 isolates, 97.3 %) and 73 isolates (52.1 %) of E. faecalis. Only four among 112 isolates of E. faecium (3.6 %) were identified as biofilm producers, including three strong producers and a single weak producer.
Table 3.
Antimicrobial susceptibility profiles of enterococcal isolates from invasive infections
| Species | MIC (mg/L) | Number of isolates (%) and 95 % confidence intervals of % Ra | |||||
|---|---|---|---|---|---|---|---|
| Antimicrobial agentb | Range | MIC50 | MIC90 | S | Ic | R | 95 % CI |
| E. faecalis (n = 140) | |||||||
| Ampicillin | 0.5–4.0 | 2.0 | 4.0 | 140 (100 %) | 0 | 0 | 0-3.3 |
| Penicillin | 0.125–16.0 | 2.0 | 4.0 | 139 (99.3 %) | - | 1 (0.7 %) | 0-4.5 |
| Vancomycin | 0.125–4.0 | 2.0 | 4.0 | 140 (100 %) | - | 0 | 0-3.3 |
| Teicoplanin | 0.125–0.5 | 0.125 | 0.25 | 140 (100 %) | - | 0 | 0-3.3 |
| HLGR | 4.0– >1024 | 1024 | >1024 | 62 (44.29 %) | - | 78 (55.71 %) | 47.1-64.0 |
| HLSR | 16.0– >2048 | 2048 | >2048 | 64 (45.71 %) | - | 76 (54.29 %) | 45.7-62.7 |
| Tetracycline | 0.125– >256 | 128 | 256 | 14 (10 %) | 2 (1.43 %) | 124 (88.57 %) | 81.9-93.1 |
| Tigecycline | 0.015–0.25 | 0.062 | 0.25 | 140 (100 %) | 0 | 0 | 0-3.3 |
| Chloramphenicol | 0.5–128 | 8.0 | 128 | 97 (69.29 %) | 1 (0.71 %) | 42 (30 %) | 22.7-38.4 |
| Daptomycin | 0.125–4.0 | 1.0 | 2.0 | 140 (100 %) | - | 0 | 0-3.3 |
| Rifampin | 0.25–16.0 | 2.0 | 8.0 | 45 (32.14 %) | 37 (26.43 %) | 58 (41.43 %) | 32.6-49.3 |
| Ciprofloxacin | 0.125– >32.0 | 8.0 | >32 | 56 (40 %) | 13 (9.29 %) | 71 (50.71 %) | 41.5-58.5 |
| Linezolid | 0.25–4.0 | 2.0 | 4.0 | 140 (100 %) | - | 0 | 0-3.3 |
| Imipenem | 0.5-64 | 16 | 64 | 11 (7.9 %) | 30 (21.4 %) | 99 (70.7 %) | 62.3-77.9 |
| Trimethoprim-sulphamethoxazole | 0.015-32 | 4 | 32 | 11 (7.9 %) | 47 (33.8 %) | 81 (58.3 %) | 49.6-66.5 |
| E. faecium (n = 112) | |||||||
| Ampicillin | 1– >256 | 64 | 128 | 1 (0.89 %) | 11 (9.82 %) | 110 (89.29 %) | 81.7-94.1 |
| Penicillin | 0.25– >256 | 128 | 256 | 9 (8.04 %) | - | 103 (91.96 %) | 84.9-96.1 |
| Vancomycin | 0.5– >256 | 2.0 | 4.0 | 104 (92.86 %) | - | 8 (7.14 %) | 3.3-14.0 |
| Teicoplanin | 0.125–128 | 0.25 | 1.0 | 107 (95.54 %) | - | 5 (4.46 %) | 1.7-10.7 |
| HLGR | 4.0– >1024 | >1024 | >1024 | 15 (13.39 %) | - | 97 (86.61 %) | 78.6-92.1 |
| HLSR | 4.0– >2048 | 2048 | >2048 | 16 (14.29 %) | - | 96 (85.71 %) | 77.5-91.4 |
| Tetracycline | 0.25–256 | 16 | 128 | 49 (43.75 %) | 4 (3.57 %) | 59 (52.68 %) | 43.1-62.1 |
| Tigecycline | 0.015–0.5 | 0.062 | 0.25 | 111 (99.11 %) | 1 (0.89 %) | 0 | 0-4.1 |
| Chloramphenicol | 0.062–32.0 | 8.0 | 16 | 89 (79.46 %) | 16 (14.29 %) | 7 (6.25 %) | 2.7-12.8 |
| Daptomycin | 0.062–4.0 | 2.0 | 4.0 | 112 (100 %) | 0 | 0 | 0.0-4.1 |
| Rifampin | 0.062– >128 | 32 | >128 | 5 (4.46 %) | 3 (2.68 %) | 104 (92.86 %) | 86-96.7 |
| Ciprofloxacin | 2.0– >256 | 256 | >256 | 0 | 1 (0.89 %) | 111 (99.11 %) | 94.4-100 |
| Linezolid | 1.0–4.0 | 2.0 | 4.0 | 112 (100 %) | 0 | 0 | 0.0-4.1 |
| Quinupristin-dalfopristin | 0.125–16 | 2.0 | 4.0 | 54 (48.2 %) | 52 (46.4 %) | 6 (%54 %) | 2.2-11.8 |
| Other Enterococcus spp. (n = 7) | |||||||
| Ampicillin | 0.0625–128.0 | 16 | 128 | 3 | 0 | 4 | - |
| Penicillin | 0.5–64.0 | 8 | 64 | 6 | - | 1 | - |
| Vancomycin | 0.5–8.0 | 2 | 8 | 4 | - | 3 | - |
| Teicoplanin | 0.125–1.0 | 0.25 | 1 | 7 | - | 0 | - |
| HLGR | 4.0–1024 | 64 | 1024 | 5 | - | 2 | - |
| HLSR | 32.0–2048 | 256 | 2048 | 4 | - | 3 | - |
| Tetracycline | 0.125–64 | 64 | 64 | 2 | 0 | 5 | - |
| Tigecycline | 0.031–0.062 | 0.031 | 0.062 | 7 | 0 | 0 | - |
| Chloramphenicol | 0.25–8.0 | 4 | 8 | 7 | 0 | 0 | - |
| Daptomycin | 0.25–2.0 | 1 | 2 | 7 | 0 | 0 | - |
| Rifampin | 0.062–64.0 | 0.5 | 64 | 5 | 0 | 2 | - |
| Ciprofloxacin | 0.125–64.0 | 4 | 64 | 3 | 0 | 4 | - |
| Linezolid | 0.015–4.0 | 2 | 4 | 7 | 0 | 0 | - |
aS, susceptible; I, intermediate; R, resistant; bHLGR, high-level gentamicin resistance, HLSR, high-level streptomycin resistance; c -, intermediate category not defined for this compound
Molecular typing of E. faecium isolates and the distribution meroclone- and lineage-specific markers
MLVA revealed 12 MLVA types (MTs; Table 4) among 112 invasive E. faecium isolates, with the most prevalent being MT159, characteristic for 52 isolates (43.3 %) from 20 hospitals. Other frequently encountered MTs included 1, 11, and 12. The subsequent MLST analysis, performed for a group of 46 representative isolates, revealed the presence of 14 STs (Table 4). The comparative eBURST analysis included four STs (78, 192, 341, 412) in lineage 78 and nine STs (17, 18, 64, 80, 117, 202, 262, 877, 878) in lineage 17/18; the remaining ST879 represented a singleton. Isolates of lineage 17/18 were most often associated with MT1 and MT12, and isolates of lineage 78 typically had MT159, however, one ST117 isolate (lineage 17/18) had MT159 (verified by repeated typing). The acquisition of vancomycin-resistance determinants vanA and vanB occurred chiefly among isolates of lineage 17/18 (five out of seven cases). Most of the isolates able to produce biofilm (three out of four) also belonged to this lineage. IS16 and espEfm were detected in all E. faecium isolates, and intAICEEfm1 for 107 isolates (95.5 %). Thus, the espEfm gene (verified by sequencing) was present in five isolates negative for intAICEEfm1. Distribution of genes located within six GDRs characteristic for ST203 from lineage 78 [12] was assessed among isolates of E. faecium collected in this study and for comparative purposes among 52 sewage isolates, not associated with the hospital meroclone [23]. Significant differences in the prevalence were found for GDR1, which was more frequently detected in sewage isolates, and GDR2 and GDR3, which occurred chiefly among hospital-associated isolates (Table 5). Comparison between two hospital lineages of E. faecium revealed a significant over-representation of all six GDRs in lineage 78 in comparison to lineage 17/18. Sequencing of PCR products for a few randomly-selected representatives of both lineages and non-hospital E. faecium for all six GDRs revealed their 100 % identity with the counterparts in ST203 [12].
Table 4.
MLVA and MLST of invasive E. faecium isolates
| MTa | VNTR profileb | Number of isolates | Number of hospitals | Number of VRE | Number of fatal cases | Number of isolates analysed by MLST | STsa | Lineage |
|---|---|---|---|---|---|---|---|---|
| 1 | 5-7-3-3-2-3 | 16 | 10 | 3 | 3 | 8 | 17; 18; 64; 80; 202 | 17/18 |
| 7 | 5-7-3-3-2-2 | 1 | 1 | 0 | 0 | 1 | 17 | 17/18 |
| 10 | 5-7-3-3-3-3 | 1 | 1 | 0 | 0 | 1 | 262 | 17/18 |
| 11 | 6-7-3-3-2-3 | 13 | 8 | 0 | 1 | 3 | 202 | 17/18 |
| 12 | 5-7-3-3-1-3 | 15 | 8 | 2 | 4 | 6 | 877; 117 | 17/18 |
| 112 | 3-7-4-2-1-3 | 1 | 1 | 0 | 0 | 1 | 878 | 17/18 |
| 159 | 5-7-3-3-1-2 | 1 | 1 | 0 | 0 | 1 | 117 | 17/18 |
| 520 | 2-7-3-3-1-2 | 4 | 4 | 1 | 1 | 2 | 117 | 17/18 |
| summary | 52 | 21 | 6 | 9 | 23 | 17/18 | ||
| 159 | 5-7-3-3-1-2 | 51 | 20 | 2 | 11 | 12 | 78; 192; 341; 412 | 78 |
| 291 | 5-7-4-3-1-2 | 4 | 2 | 0 | 1 | 1 | 78 | 78 |
| 334 | 5-7-3-4-1-2 | 3 | 1 | 0 | 2 | 1 | 78 | 78 |
| 518 | 5-2-3-3-1-2 | 1 | 1 | 0 | 0 | 1 | 78 | 78 |
| summary | 59 | 21 | 2 | 14 | 15 | 78 | ||
| 519 | 3-7-3-3-2-2 | 1 | 1 | 0 | 0 | 1 | 879 | singleton |
anovel MTs and STs underlined; bin the order: VNTR1, VNTR2, VNTR7, VNTR8, VNTR9, VNTR10
Table 5.
Distribution of GDRs among lineages 17/18 and 78 and among hospital and sewage isolates of E. faecium
| lineagea/origin | GDR1 (%) | GDR2 (%) | GDR3 (%) | GDR5 (%) | GDR6 (%) | GDR7 (%) | Number of isolates with n GDR loci | ||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| n = 0 | n = 1 | n = 2 | n = 3 | n = 4 | n = 5 | n = 6 | |||||||
| 17/18 (n = 27) | 14 (51.2) | 16 (59.2) | 5 (18.5) | 4 (14.8) | 6 (22.2) | 2 (7.4) | 4 | 12 | 4 | 3 | 3 | 0 | 1b |
| 78 (n = 18) | 16 (88.9) | 18 (100) | 15 (83.3) | 18 (100) | 11 (61.1) | 13 (72.2) | 0 | 0 | 0 | 2 | 3 | 5 | 8 |
| P c | 0.02 | 0.005 | 0.0001 | 0.00000 | 0.02 | 0.00003 | |||||||
| ST879 (n = 1) | 1 | 0 | 1 | 1 | 0 | 1 | 1 | ||||||
| Hospital (n = 46) | 31 (67.4) | 34 (73.9) | 21 (45.6) | 23 (50.0) | 17 (36.9) | 16 (34.8) | 4 | 12 | 4 | 5 | 7 | 5 | 9 |
| Sewage (n = 52) | 49 (94.2) | 0 (0) | 5 (9.6) | 21 (40.4) | 11 (21.1) | 11 (21.1) | 2 | 20 | 13 | 16 | 1 | 0 | 0 |
| P d | 0.001 | 0 | 0.0001 | 0.4 | 0.1 | 0.2 | |||||||
aBased on the MLST data; bverified by repeated typing and GDR sequencing; cdifferences in GDR distributions between lineages 17/18 and 78 isolates; ddifferences in GDR distributions between hospital and sewage isolates
Discussion
The growing importance of enterococcal infections in hospitals prompted us to perform an enhanced surveillance of enterococci causing invasive diseases in selected Polish hospitals in the pre-defined period of time. The clinical presentations of infections were typical for enterococci, including mainly bacteraemia and abdominal infections. Similarly to other reports, our results point out the abdominal infections as the most frequently observed sources of bacteraemia/septicaemia; however, the small proportion of E. faecalis bacteraemia originating from urinary tract (7.1 %) was surprising [24, 25]. This could be due to the relatively high ratio of bacteraemia with unknown source observed in our study for both E. faecalis and E. faecium (40.7 % and 46.4 % respectively) in comparison to other reports (E. faecalis 30 % and 21.6 %; E. faecium 39.9 % and 20.7 %) [24, 25]. Collected clinical data confirmed established risk factors for acquisition of invasive enterococcal infection, such as advanced age, previous hospitalization, and ICU stay [13, 24]. In our study, the majority of patients were male and aged above 50 years, and most of the fatal cases were observed in this age group. Other studies have reported similar proportions of patients’ gender and age distribution [24]. The 14-day mortality rate of 18.1 % reported in our study corresponded to the 30-day mortality rates of 18.9 % to 25 % among patients with enterococcal bacteraemia in other countries [25]; however, it is important to note that at the 2-week reporting interval the majority of patients in our study were still under treatment. We observed a higher mortality in infections caused by E. faecium than E. faecalis (20.1 % vs 15.4 %), which is in agreement with a report from Denmark (34.6 % and 21.4 % respectively) [24] and Spain (30 % and 26 % respectively, among cancer patients) [26].
While at the rise of enterococcal infections in hospitals in the 1970s E. faecalis represented the most predominant species, contributing to approximately 90 % of infections, more recently the relative proportion of these two species has been changing in the favour of E. faecium and is now almost reaching or even exceeding the parity [26, 27], due to the increasing incidence of infections caused by E. faecium [24]. This worldwide trend of increasing importance of E. faecium in HAIs was also observed in the present study, where 54 % and 43 % of infections were caused by E. faecalis and E. faecium respectively. For comparison, the ratio of E. faecium accounted for around 30 % of isolates from blood in our country during 2001–2004, and increased to around 40 % in 2009–2013 (http://www.ecdc.europa.eu/en/activities/surveillance/EARS-Net). This shift in the proportion of E. faecium to E. faecalis results in the increased overall morbidity of enterococcal infections, due to a higher fatality ratio of bacteraemia caused by this species [26], which may be at least partially associated with frequent resistance of E. faecium to important anti-enterococcal drugs, such as ampicillin, aminoglycosides, and glycopeptides [24–26]. Ampicillin resistance is now very common among hospital E. faecium, and it proceeded the appearance and constant rise of vancomycin resistance in the hospital meroclone of this species [11, 28]. In our study, acquired resistance to vancomycin in E. faecium was still relatively rare (7 %). VREfm have been observed in Poland since the end of the 1990s with an increasing incidence [19, 29]. The frequency of VRE differs in various regions of the world, with a high prevalence reported in the US (up to 80 % of E. faecium) [27] and some European countries, such as Greece, the UK, and Portugal [28], while a low ratio (below 1 %) is characteristic for Sweden, The Netherlands, France, and Spain (http://www.ecdc.europa.eu/en/activities/surveillance/EARS-Net). We observed a very high prevalence of high-level resistance to aminoglycosides in E. faecium, which eliminates the possibility of combined therapy including aminoglycoside with cell-wall-active agents (penicillins, glycopeptides) for synergistic bactericidal effect. Although E. faecalis remains generally susceptible to penicillins, high-level resistance to aminoglycosides exceeded 50 % in our study, with the same problem for the combined therapy. Such a phenotype has also been reported for several other European countries (http://www.ecdc.europa.eu/en/activities/surveillance/EARS-Net), and our previous analysis of nosocomial isolates of E. faecalis from six European countries showed a significant role of two hospital clones in the spread of high-level aminoglycoside resistance in this species [8]. All isolates in the current study showed susceptibility to linezolid, the drug of last resort in VRE infections, although resistance to this compound is being observed in our country for both E. faecalis and E. faecium [30].
Selection of particular enterococcal clones, adapted to hospital settings, appears to play a crucial role in the increasing importance of these pathogens in HAIs, especially in the case of E. faecium. The rise in the prevalence of this species which was found in our study prompted us to analyse collected E. faecium isolates in more detail. The results of MLVA, MLST and detection of markers, specific for nosocomial E. faecium such as IS16 [18], espEfm and intAICEEfm1 associated with a transferable pathogenicity island ICEfm1 [31, 32], indicated that almost all isolates represented hospital epidemic meroclone. Circulation of strains belonging to this clone most likely predates the appearance of the first VRE in Polish hospitals in the late 1990s [19, 29]. EspEfm, a species-specific variant of the enterococcal surface protein, is involved in biofilm formation [33] and increases enterococcal virulence in endocarditis [34]. Although the espEfm gene was present ubiquitously in the studied group, only a minority of isolates were able to form biofilm under experimental conditions. A recent study [35] questioned the utility of a polystyrene dish assay for biofilm formation by E. faecalis, and showed much more reliable performance of porcine heart valve explants. A similar system may also be required for E. faecium biofilm studies.
Hospital E. faecium in our study belonged to both major BAPS groups, corresponding to lineages 17/18 and 78 [10]. The latter, more recently evolving lineage is currently being isolated in hospitals all over the world [26, 36, 37], and in Poland it was first observed in 2005 among VREfm [19]. ST78 and its variants are typically associated with MT159 [38, 39], the most prevalent MT in our study. Spread of lineage 78 strains is considered to be a significant factor of the increasing VanB-type vancomycin resistance in Australia [37], and current detailed genomic analysis of strains representing both lineages has revealed the presence of seven GDRs, additionally present in a representative of lineage 78 [12]. In this study, we analysed the distribution of genes characteristic for six GDRs, with the exception of one region (GDR4) which contains a presumable integrase gene. Genes of two GDRs, namely the gene encoding a riboflavin biosynthesis protein RibD from GDR2 and the gene encoding a putative phosphosugar isomerase from GDR3, were significantly more prevalent among isolates from hospital settings in comparison to sewage isolates, while the latter were more often carrying the gene of cadmium-translocating P-type ATPase from GDR1; it is likely that the product of this gene provides a selective advantage in various environments. Among hospital strains, all six analysed GDRs were over-represented among successful lineage 78; however, genes of GDR5, 6, and 7 were relatively frequent also among the sewage isolates, suggesting that E. faecium circulating in the community might have been a source of such genes for nosocomial strains. Further analyses are, however, required to verify such a possibility.
In summary, our study provides the data on species distribution, prevalence of resistance, and clonality of enterococcal invasive isolates, and characterizes patients affected by such infections. The study on Polish isolates shows the similarity of invasive, hospital-adapted E. faecium to strains circulating in other countries, and underlines the importance of permanent surveillance of the dynamic epidemiological situation concerning these dangerous opportunistic pathogens. These data will be the reference for future studies performed in Poland.
Acknowledgments
We thank the representatives of all the centres participating in the study: Krzysztof Golec, Anna Onyszk, Katarzyna Muszyńska, Elżbieta Maćkowiak, Jolanta Zielińska, Małgorzata Pilarska, Małgorzata Szarata, Krzysztof Burdynowski, Halina Kondracka-Grabarczyk, Grzegorz Ziółkowski, Aleksandra Renik, Beata Klonowska, Dorota Kociszewska, Anna Jędrzejewska, Joanna Humanowska, Małgorzata Kozak, Maria Szymankiewicz, Jolanta Prawda-Zołotar, Elżbieta Iskra, Anna Mól, Bonita Durnaś, Mirosława Piotrowska, Zofia Żak-Puławska, Wojciech Żulikowski, Anna Budzyńska, Julianna Kurlenda, Anita Mądroszyk, Edyta Podsiadły, and Jolanta Szumilewicz for providing the isolates, and Janetta Top for assigning new alleles and STs, and for her aid in assigning MTs. This publication made use of the Enterococcus faecium MLST websites http://efaecium.mlst.net/ hosted at Imperial College and developed and funded by the Wellcome Trust, and http://pubmlst.org/efaecium/ sited at the University of Oxford and funded by the Wellcome Trust. Parts of this publication have previously been presented at the 22nd European Congress of Clinical Microbiology and Infectious Diseases (ECCMID), 31 March–3 April 2012, London, UK (oral presentation).
Compliance with ethical standards
Funding
This study was partially financed by health programme “Narodowy Program Ochrony Antybiotyków na lata 2011-2015” from The Polish Ministry of Health and by the grant SPUB MIKROBANK from the Polish Ministry of Science and Higher Education.
Conflict of interest
The authors declare that they have no conflict of interest.
Ethical approval
The authors declare that ethical approval was not required.
Informed consent
The authors declare that informed consent was not required.
Footnotes
I. Gawryszewska and D. Żabicka contributed equally to this work.
References
- 1.Wisplinghoff H, Bischoff T, Tallent SM, Seifert H, Wenzel RP, Edmond MB. Nosocomial bloodstream infections in US hospitals: analysis of 24,179 cases from a prospective nationwide surveillance study. Clin Infect Dis. 2004;39(3):309–317. doi: 10.1086/421946. [DOI] [PubMed] [Google Scholar]
- 2.European Centre for Disease Prevention and Control (2013) Point prevalence survey of healthcare-associated infections and antimicrobial use in European acute care hospitals. ECDC, Stockholm. doi:10.2900/86011
- 3.Hegstad K, Mikalsen T, Coque TM, Werner G, Sundsfjord A. Mobile genetic elements and their contribution to the emergence of antimicrobial resistant Enterococcus faecalis and Enterococcus faecium. Clin Microbiol Infect. 2010;16(6):541–554. doi: 10.1111/j.1469-0691.2010.03226.x. [DOI] [PubMed] [Google Scholar]
- 4.Gordon S, Swenson JM, Hill BC, Pigott NE, Facklam RR, Cooksey RC, Thornsberry C, Jarvis WR, Tenover FC. Antimicrobial susceptibility patterns of common and unusual species of enterococci causing infections in the United States. Enterococcal Study Group. J Clin Microbiol. 1992;30(9):2373–2378. doi: 10.1128/jcm.30.9.2373-2378.1992. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Homan WL, Tribe D, Poznanski S, Li M, Hogg G, Spalburg E, Van Embden JD, Willems RJ. Multilocus sequence typing scheme for Enterococcus faecium. J Clin Microbiol. 2002;40(6):1963–1971. doi: 10.1128/JCM.40.6.1963-1971.2002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Ruiz-Garbajosa P, Bonten MJ, Robinson DA, Top J, Nallapareddy SR, Torres C, Coque TM, Cantón R, Baquero F, Murray BE, del Campo R, Willems RJ. Multilocus sequence typing scheme for Enterococcus faecalis reveals hospital-adapted genetic complexes in a background of high rates of recombination. J Clin Microbiol. 2006;44(6):2220–2228. doi: 10.1128/JCM.02596-05. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Paulsen IT, Banerjei L, Myers GS, Nelson KE, Seshadri R, Read TD, Fouts DE, Eisen JA, Gill SR, Heidelberg JF, Tettelin H, Dodson RJ, Umayam L, Brinkac L, Beanan M, Daugherty S, DeBoy RT, Durkin S, Kolonay J, Madupu R, Nelson W, Vamathevan J, Tran B, Upton J, Hansen T, Shetty J, Khouri H, Utterback T, Radune D, Ketchum KA, Dougherty BA, Fraser CM. Role of mobile DNA in the evolution of vancomycin-resistant Enterococcus faecalis. Science. 2003;299(5615):2071–2074. doi: 10.1126/science.1080613. [DOI] [PubMed] [Google Scholar]
- 8.Kuch A, Willems RJ, Werner G, Coque TM, Hammerum AM, Sundsfjord A, Klare I, Ruiz-Garbajosa P, Simonsen GS, van Luit-Asbroek M, Hryniewicz W, Sadowy E. Insight into antimicrobial susceptibility and population structure of contemporary human Enterococcus faecalis isolates from Europe. J Antimicrob Chemother. 2012;67(3):551–558. doi: 10.1093/jac/dkr544. [DOI] [PubMed] [Google Scholar]
- 9.Willems RJ, Top J, van Santen M, Robinson DA, Coque TM, Baquero F, Grundmann H, Bonten MJ. Global spread of vancomycin-resistant Enterococcus faecium from distinct nosocomial genetic complex. Emerg Infect Dis. 2005;11(6):821–828. doi: 10.3201/1106.041204. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Willems RJ, Top J, van Schaik W, Leavis H, Bonten M, Sirén J, Hanage WP, Corander J. Restricted gene flow among hospital subpopulations of Enterococcus faecium. Ambio. 2012;3(4):e00151–12. doi: 10.1128/mBio.00151-12. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Top J, Willems R, Bonten M. Emergence of CC17 Enterococcus faecium: from commensal to hospital-adapted pathogen. FEMS Immunol Med Microbiol. 2008;52(3):297–308. doi: 10.1111/j.1574-695X.2008.00383.x. [DOI] [PubMed] [Google Scholar]
- 12.Lam MM, Seemann T, Tobias NJ, Chen H, Haring V, Moore RJ, Ballard S, Grayson LM, Johnson PD, Howden BP, Stinear TP. Comparative analysis of the complete genome of an epidemic hospital sequence type 203 clone of vancomycin-resistant Enterococcus faecium. BMC Genomics. 2013;14:595. doi: 10.1186/1471-2164-14-595. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Billington EO, Phang SH, Gregson DB, Pitout JD, Ross T, Church DL, Laupland KB, Parkins MD. Incidence, risk factors, and outcomes for Enterococcus spp. blood stream infections: a population-based study. Int J Infect Dis. 2014;26:76–82. doi: 10.1016/j.ijid.2014.02.012. [DOI] [PubMed] [Google Scholar]
- 14.Magiorakos AP, Srinivasan A, Carey RB, Carmeli Y, Falagas ME, Giske CG, Harbarth S, Hindler JF, Kahlmeter G, Olsson-Liljequist B, Paterson DL, Rice LB, Stelling J, Struelens MJ, Vatopoulos A, Weber JT, Monnet DL. Multidrug-resistant, extensively drug-resistant and pandrug-resistant bacteria: an international expert proposal for interim standard definitions for acquired resistance. Clin Microbiol Infect. 2012;18(3):268–281. doi: 10.1111/j.1469-0691.2011.03570.x. [DOI] [PubMed] [Google Scholar]
- 15.Tsikrikonis G, Maniatis AN, Labrou M, Ntokou E, Michail G, Daponte A, Stathopoulos C, Tsakris A, Pournaras S. Differences in biofilm formation and virulence factors between clinical and fecal enterococcal isolates of human and animal origin. Microb Pathog. 2012;52(6):336–343. doi: 10.1016/j.micpath.2012.03.003. [DOI] [PubMed] [Google Scholar]
- 16.Baldassarri L, Cecchini R, Bertuccini L, Ammendolia MG, Iosi F, Arciola CR, Montanaro L, Di Rosa R, Gherardi G, Dicuonzo G, Orefici G, Creti R. Enterococcus spp. produces slime and survives in rat peritoneal macrophages. Med Microbiol Immunol. 2001;190(3):113–120. doi: 10.1007/s00430-001-0096-8. [DOI] [PubMed] [Google Scholar]
- 17.Shankar V, Baghdayan AS, Huycke MM, Lindahl G, Gilmore MS. Infection-derived Enterococcus faecalis strains are enriched in esp, a gene encoding a novel surface protein. Infect Immun. 1999;67(1):193–200. doi: 10.1128/iai.67.1.193-200.1999. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Werner G, Fleige C, Geringer U, van Schaik W, Klare I, Witte W. IS element IS16 as a molecular screening tool to identify hospital-associated strains of Enterococcus faecium. BMC Infect Dis. 2011;11:80. doi: 10.1186/1471-2334-11-80. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Sadowy E, Sieńko A, Gawryszewska I, Bojarska A, Malinowska K, Hryniewicz W. High abundance and diversity of antimicrobial resistance determinants among early vancomycin-resistant Enterococcus faecium in Poland. Eur J Clin Microbiol Infect Dis. 2013;32(9):1193–1203. doi: 10.1007/s10096-013-1868-y. [DOI] [PubMed] [Google Scholar]
- 20.Clark NC, Cooksey RC, Hill BC, Swenson JM, Tenover FC. Characterization of glycopeptide-resistant enterococci from U.S. hospitals. Antimicrob Agents Chemother. 1993;37(11):2311–2317. doi: 10.1128/AAC.37.11.2311. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Dahl KH, Simonsen GS, Olsvik O, Sundsfjord A. Heterogeneity in the vanB gene cluster of genomically diverse clinical strains of vancomycin-resistant enterococci. Antimicrob Agents Chemother. 1999;43(5):1105–1110. doi: 10.1128/aac.43.5.1105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Top J, Schouls LM, Bonten MJ, Willems RJ. Multiple-locus variable-number tandem repeat analysis, a novel typing scheme to study the genetic relatedness and epidemiology of Enterococcus faecium isolates. J Clin Microbiol. 2004;42(10):4503–4511. doi: 10.1128/JCM.42.10.4503-4511.2004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Sadowy E, Luczkiewicz A. Drug-resistant and hospital-associated Enterococcus faecium from wastewater, riverine estuary and anthropogenically impacted marine catchment basin. BMC Microbiol. 2014;14:66. doi: 10.1186/1471-2180-14-66. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Pinholt M, Ostergaard C, Arpi M, Bruun NE, Schønheyder HC, Gradel KO, Søgaard M, Knudsen JD, Danish Collaborative Bacteraemia Network (DACOBAN) Incidence, clinical characteristics and 30-day mortality of enterococcal bacteraemia in Denmark 2006–2009: a population-based cohort study. Clin Microbiol Infect. 2014;20(2):145–151. doi: 10.1111/1469-0691.12236. [DOI] [PubMed] [Google Scholar]
- 25.McBride SJ, Upton A, Roberts SA. Clinical characteristics and outcomes of patients with vancomycin-susceptible Enterococcus faecalis and Enterococcus faecium bacteraemia-a five-year retrospective review. Eur J Clin Microbiol Infect Dis. 2010;29(1):107–114. doi: 10.1007/s10096-009-0830-5. [DOI] [PubMed] [Google Scholar]
- 26.Gudiol C, Ayats J, Camoez M, Domínguez MÁ, García-Vidal C, Bodro M, Ardanuy C, Obed M, Arnan M, Antonio M, Carratalà J. Increase in bloodstream infection due to vancomycin-susceptible Enterococcus faecium in cancer patients: risk factors, molecular epidemiology and outcomes. PLoS One. 2013;8(9):e74734. doi: 10.1371/journal.pone.0074734. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Hidron AI, Edwards JR, Patel J, Horan TC, Sievert DM, Pollock DA, Fridkin SK, National Healthcare Safety Network Team; Participating National Healthcare Safety Network Facilities (2008) NHSN annual update: antimicrobial-resistant pathogens associated with healthcare-associated infections: annual summary of data reported to the National Healthcare Safety Network at the Centers for Disease Control and Prevention, 2006–2007. Infect Control Hosp Epidemiol 29(11):996–1011, Erratum in: Infect Control Hosp Epidemiol 30(1):107. doi:10.1086/595835 [DOI] [PubMed]
- 28.Werner G, Coque TM, Hammerum AM, Hope R, Hryniewicz W, Johnson A, Klare I, Kristinsson KG, Leclercq R, Lester CH, Lillie M, Novais C, Olsson-Liljequist B, Peixe LV, Sadowy E, Simonsen GS, Top J, Vuopio-Varkila J, Willems RJ, Witte W, Woodford N (2008) Emergence and spread of vancomycin resistance among enterococci in Europe. Euro Surveill 13(47). http://www.eurosurveillance.org/ViewArticle.aspx?ArticleId=19046 [PubMed]
- 29.Hryniewicz W, Szczypa K, Bronk M, Samet A, Hellman A, Trzcinski K. First report of vanconmycin-resistant Enterococcus faecium isolated in Poland. Clin Microbiol Infect. 1999;5(8):503–505. doi: 10.1111/j.1469-0691.1999.tb00181.x. [DOI] [PubMed] [Google Scholar]
- 30.Gawryszewska I, Żabicka D, Hryniewicz W, Sadowy E (2014) Linezolid resistant enterococci – resistance mechanisms and clonal relatedness. 24th ECCMID 2014. http://www.escmid.org/escmid_library/online_lecture_library
- 31.Willems RJ, Homan W, Top J, van Santen-Verheuvel M, Tribe D, Manzioros X, Gaillard C, Vandenbroucke-Grauls CM, Mascini EM, van Kregten E, van Embden JD, Bonten MJ. Variant esp gene as a marker of a distinct genetic lineage of vancomycin-resistant Enterococcus faecium spreading in hospitals. Lancet. 2001;357(9259):853–855. doi: 10.1016/S0140-6736(00)04205-7. [DOI] [PubMed] [Google Scholar]
- 32.Top J, Sinnige JC, Majoor EA, Bonten MJ, Willems RJ, van Schaik W. The recombinase IntA is required for excision of esp-containing ICEEfm1 in Enterococcus faecium. J Bacteriol. 2011;193(4):1003–1006. doi: 10.1128/JB.00952-10. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Heikens E, Bonten MJ, Willems RJ. Enterococcal surface protein Esp is important for biofilm formation of Enterococcus faecium E1162. J Bacteriol. 2007;189(22):8233–8240. doi: 10.1128/JB.01205-07. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Heikens E, Singh KV, Jacques-Palaz KD, van Luit-Asbroek M, Oostdijk EA, Bonten MJ, Murray BE, Willems RJ. Contribution of the enterococcal surface protein Esp to pathogenesis of Enterococcus faecium endocarditis. Microbes Infect. 2011;13(14–15):1185–1890. doi: 10.1016/j.micinf.2011.08.006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Leuck AM, Johnson JR, Dunny GM. A widely used in vitro biofilm assay has questionable clinical significance for enterococcal endocarditis. PLoS One. 2014;9(9):e107282. doi: 10.1371/journal.pone.0107282. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Hsieh YC, Lee WS, Ou TY, Hsueh PR. Clonal spread of CC17 vancomycin-resistant Enterococcus faecium with multilocus sequence type 78 (ST78) and a novel ST444 in Taiwan. Eur J Clin Microbiol Infect Dis. 2010;29(1):25–30. doi: 10.1007/s10096-009-0810-9. [DOI] [PubMed] [Google Scholar]
- 37.Wardal E, Markowska K, Zabicka D, Wróblewska M, Giemza M, Mik E, Połowniak-Pracka H, Woźniak A, Hryniewicz W, Sadowy E. Molecular analysis of vanA outbreak of Enterococcus faecium in two Warsaw hospitals: the importance of mobile genetic elements. Biomed Res Int. 2014;2014:575367. doi: 10.1155/2014/575367. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Coombs GW, Pearson JC, Daley DA, Le T, Robinson OJ, Gottlieb T, Howden BP, Johnson PD, Bennett CM, Stinear TP, Turnidge JD, Australian Group on Antimicrobial Resistance Molecular epidemiology of enterococcal bacteremia in Australia. J Clin Microbiol. 2014;52(3):897–905. doi: 10.1128/JCM.03286-13. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Werner G, Klare I, Witte W. The current MLVA typing scheme for Enterococcus faecium is less discriminatory than MLST and PFGE for epidemic-virulent, hospital-adapted clonal types. BMC Microbiol. 2007;7:28. doi: 10.1186/1471-2180-7-28. [DOI] [PMC free article] [PubMed] [Google Scholar]