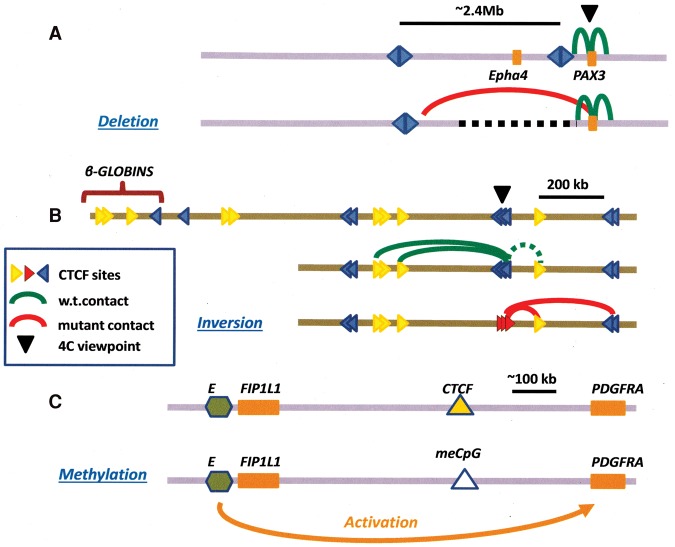
Figure 2.
Effects of altering CTCF-binding sites on domain structure and gene expression. (A) Effect on 4C contacts of deleting DNA containing an insulator boundary near the mouse PAX3 gene, showing novel interactions with regions further upstream (Lupianez et al. 2015). Disruption of a TAD boundary had been shown earlier to cause ectopic chromosomal contacts and long-range transcriptional misregulation in the mouse Xist locus (Nora et al. 2012; see also Dowen et al. 2014). (B) Effect of inverting CTCF-binding sites on the pattern of 4C contacts near the mouse β-globin locus. The dotted green interaction line calls attention to the nonconvergent orientation of the CTCF sites marked by the blue triangles and the yellow one immediately downstream. After inversion, contacts between the red (inverted) sites and the yellow site actually strengthened despite the fact that the sites are not facing toward each other on the loop (Guo et al. 2015; see also de Wit et al. 2015) (C) Effect of methylation of a CTCF site on boundary activity. In certain human gliomas, the product of the mutated isocitrate dehydrogenase (IDH) gene interferes with DNA demethylation at a critical CTCF-binding site, resulting in loss of CTCF binding and insulation and inappropriate activation of the PDGFRA gene, a glioma oncogene, by a distal enhancer (green hexagon) (Flavahan et al. 2016).