Here, Carmona et al. show that two ancestral RAG1 proteins, Transib transposase and purple sea urchin RAG1-like, have a latent ability to initiate V(D)J recombination when coexpressed with RAG2 and that in vitro transposition by Transib transposase is stimulated by RAG2. They propose that evolution of RAG1/RAG2 began with a Transib transposon whose intrinsic recombination activity was enhanced by capture of an ancestral RAG2, allowing for the development of adaptive immunity.
Keywords: RAG, Transib, V(D)J recombination, evolution, transposition
Abstract
The recombination-activating gene 1 (RAG1) and RAG2 proteins initiate V(D)J recombination, the process that assembles the B- and T-lymphocyte antigen receptor genes of jawed vertebrates. RAG1 and RAG2 are thought to have arisen from a transposable element, but the origins of this element are not understood. We show that two ancestral RAG1 proteins, Transib transposase and purple sea urchin RAG1-like, have a latent ability to initiate V(D)J recombination when coexpressed with RAG2 and that in vitro transposition by Transib transposase is stimulated by RAG2. Conversely, we report low levels of V(D)J recombination by RAG1 in the absence of RAG2. Recombination by RAG1 alone differs from canonical V(D)J recombination in having lost the requirement for asymmetric DNA substrates, implicating RAG2 in the origins of the “12/23 rule,” a fundamental regulatory feature of the reaction. We propose that evolution of RAG1/RAG2 began with a Transib transposon whose intrinsic recombination activity was enhanced by capture of an ancestral RAG2, allowing for the development of adaptive immunity.
V(D)J recombination is the process by which the antigen receptor genes of jawed vertebrates are assembled. By joining different combinations of variable (V), diversity (D), and joining (J) gene segments, V(D)J recombination can generate a wide range of receptor specificities, providing the molecular basis for adaptive immunity. The proteins at the heart of this reaction are encoded by recombination-activating gene 1 (RAG1) and RAG2 (Schatz et al. 1989; Oettinger et al. 1990). The RAG1 and RAG2 proteins bind to recombination signal sequences (RSSs) that demarcate the gene segments of the antigen receptor loci and target dsDNA cleavage by RAG. Subsequent repair by the nonhomologous end-joining DNA repair pathway joins the gene segments to form coding joints and the cleaved RSSs to form signal joints (Rooney et al. 2004). The RSS consists of three sequence elements: the nonamer, the spacer, and the heptamer. While the nonamer and the heptamer sequences are well conserved, the length of the spacer, either 12 or 23 base pairs, is its defining characteristic (Ramsden et al. 1994). Therefore, two types of RSS exist depending on spacer length, the 12RSS and the 23RSS, and recombination is only efficient when one 12RSS and one 23RSS are engaged by RAG, a restriction known as the 12/23 rule.
Because RAG1-deficient and RAG2-deficient mice and humans exhibit identical phenotypes characterized by a complete absence of V(D)J recombination and lymphocyte development (Mombaerts et al. 1992; Shinkai et al. 1992; Notarangelo et al. 1999), the two RAG proteins are each presumed to be essential for V(D)J recombination. Despite this, RAG1 and RAG2 play vastly different roles in the DNA cleavage reaction. RAG1 contains the RNaseH fold catalytic domain and regions that make direct contact with the RSS and is responsible for the enzymatic activity of the RAG complex (Schatz and Swanson 2011; Kim et al. 2015; Ru et al. 2015). In contrast, RAG2 interacts with and enhances the DNA-binding and cleavage functions of RAG1. RAG2 also contains a plant homeodomain (PHD) that interacts with histone H3 in which Lys4 is trimethylated (H3K4me3) and is responsible for the promiscuous association of RAG2 with transcriptionally active chromatin (Liu et al. 2007; Matthews et al. 2007; Ji et al. 2010). Therefore, RAG1 appears to contain all of the essential domains and activities needed to bind and cleave DNA, placing RAG2 in the role of an accessory or regulatory factor. Why the enzymatic activity provided by RAG1 would require regulation by an additional protein is not immediately evident and is further complicated by RAG2's contribution to the broad localization of RAG1 in the genome to sites where DNA cleavage would be detrimental (Ji et al. 2010; Teng et al. 2015).
Models for the evolution of adaptive immunity and V(D)J recombination have long postulated the origin of the RAGs as components of a transposable element (TE) (Thompson 1995; Fugmann 2010). The RSSs have a function similar to that of the terminal inverted repeats (TIRs) of cut and paste transposons, and RAG cleaves DNA by a nick-hairpin mechanism very similar to that used by several transposases (Jones and Gellert 2004). The link between RAG and transposases was strengthened by the demonstration that RAG is able to perform cut and paste DNA transposition in vitro (Agrawal et al. 1998; Hiom et al. 1998), a finding that added impetus to the search for the origin of the RAGs. Of particular interest among TEs is the widely dispersed Transib family (Kapitonov and Jurka 2005), whose TIRs resemble the RSS, particularly the heptamer, and whose transposase shares sequence similarity with RAG1 and cleaves DNA by a nick-hairpin reaction mechanism similar to that of RAG (Hencken et al. 2012). However, while this provides a candidate ancestral protein for RAG1, it does not account for RAG2, as Transib elements contain no RAG2-like entity. Discerning the origin of the RAGs has been further complicated by the identification of RAG-like genes in the genomes of animals that predate jawed vertebrates (Kapitonov and Jurka 2005; Fugmann et al. 2006; Fugmann 2010; Zhang et al. 2014; Kapitonov and Koonin 2015). In most instances, a RAG2-like gene has been identified alongside its RAG1-like partner, but in no such case has any catalytic or transposase activity been demonstrated. Therefore, the origin of RAG2 remains elusive.
Here, we propose that RAG2 played a critical role in the establishment and evolution of V(D)J recombination. We show that RAG1 can mediate low levels of recombination in the absence of RAG2 and that this activity is less dependent on the 12/23 rule than the activity mediated by RAG1 and RAG2. We report the first functional characterization of V(D)J recombination mediated by two proposed ancient RAG1 relatives and note that they exhibit activity only when paired with a vertebrate RAG2. Furthermore, we show that while Transib transposase is capable of mediating transposition by itself, its activity on its own TIRs and on RSSs is enhanced by RAG2. These findings provide the basis for a model in which RAG1 originated from a TE of the Transib family, which acquired a RAG2-like element, thereby providing functional advantages that allowed for the evolution of the adaptive immune system of jawed vertebrates.
Results
RAG1 mediates V(D)J recombination in the absence of RAG2
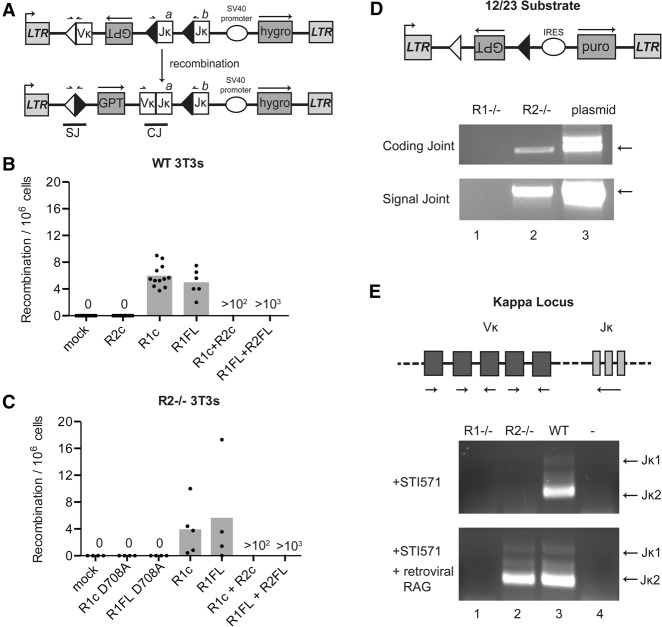
The ability of RAG1 to bind and cleave DNA, along with the existence of TEs with homology with RAG1 but without a RAG2-like counterpart (Kapitonov and Jurka 2005) and a previous report that suggested RAG2-independent activity (Montaudouin et al. 2010), led us to postulate that RAG1 functioned alone at some point in its evolutionary history and might still retain some of this activity. To test this, a previously reported sensitive assay for V(D)J recombination (Schatz and Baltimore 1988) was established in 3T3 fibroblast lines. The integrated inversional recombination substrate ZGR consists of the xanthine–guanine phosphoribosyltransferase (GPT) gene in an orientation inverse to transcription driven by the long terminal repeat (LTR) and flanked by a V segment with its endogenous 12RSS on one side and two copies of a J segment with their endogenous 23RSSs on the other side, providing two potential partners for the 12RSS (Fig. 1A). Due to the relative orientation of the RSSs, the intervening sequence is inverted upon successful recombination, allowing for expression of GPT and thereby conferring mycophenolic acid (MPA) resistance on the cell. Wild-type or RAG2−/− fibroblast lines containing this substrate were transduced to express RAG1, RAG2, or both or with a control empty vector and, after allowing time for recombination to occur, were placed in MPA selection. The minimal portion of each RAG that can mediate substantial levels of V(D)J recombination has been defined as the “core” and has been used extensively in the biochemical characterization of these proteins (Swanson 2004). However, because these truncation mutants are less active in vivo than the full-length proteins (Schatz and Swanson 2011), both core and full-length versions were tested for activity.
Figure 1.
V(D)J recombination mediated by RAG1 in the absence of RAG2. (A) Schematic of the ZGR recombination substrate. The GPT gene is inverted by recombination between the Vκ 12RSS (white) and one of the Jκ 23RSSs (black), yielding a signal joint (SJ) and coding joint (CJ) and allowing for GPT expression from the 5′ LTR and MPA resistance. (hygro) Hygromycin resistance gene; (small black arrows) coding joint and signal joint PCR primer-binding sites. (B,C) Quantitation of recombination in 3T3 fibroblast lines. Recombination efficiency for cells expressing the indicated protein or proteins plotted as the number of MPA-resistant colonies per million cells plated, with each dot indicating an independent experiment and the bar height indicating the mean. (Mock) Empty vector control; (c) core RAG proteins; (FL) full-length RAG proteins; (R1cD) D708A active site mutant of the RAG1 core; (R1flD) D708A active site mutant of RAG1 full length. (B) Wild-type 3T3 line. (C) RAG2−/− 3T3 line. (D) Substrate recombination in v-abl cells. (Top) Schematic of a simplified 12/23 inversional substrate. (Bottom) PCR assays for coding or signal joints in genomic DNA from RAG1−/− or RAG2−/− v-abl cells incubated with STI-571 and expressing RAG1 (in RAG2−/−) or RAG2 (in RAG1−/−) from retroviral vectors, representative of at least three experiments. (Plasmid) Reaction spiked with positive control template DNA. (E) Igκ locus recombination in v-abl cells. (Top) Schematic of the Igκ locus. (Bottom) Representative PCR assays for V-to-Jκ1 and V-to-Jκ2 recombination in RAG1−/− or RAG2−/− cells (expressing retroviral RAG2 or RAG1, respectively) or wild-type cells incubated with ST1–571. (Lane 4) No template control. Expected product sizes for V to Jκ1 or V to Jκ2 are indicated.
As expected (Oettinger et al. 1990), cells expressing both mouse RAG1 and RAG2, either full-length or core, yielded hundreds to thousands of recombination events per million cells screened (Fig. 1B). Strikingly, wild-type 3T3 cells expressing core or full-length mouse RAG1 also yielded a small but consistent number of recombination events (Fig. 1B). Similar results were obtained in a RAG2−/− 3T3 line, thereby ruling out any contribution from endogenous RAG2 (Fig. 1C). The identity of these events was verified through PCR assays that detect the coding and signal joints generated in the inversional recombination reaction (Supplemental Fig. S1A). Sequencing of the PCR products revealed normal signal joints and coding joints that, while normal in most regards, exhibited some alterations in the levels of insertions and deletions relative to coding joints formed in the presence of RAG2 (Supplemental Fig. S2A,B; Supplemental Table S1A,B). Importantly, expression of core or full-length RAG1 containing a D708A active site mutation (R1c D708A and R1FL D708A, respectively) did not yield any MPA-resistant colonies (Fig. 1C), and events were not detected in cells transduced with the empty vector or a vector expressing RAG2 (Fig. 1B,C). Hence, the rare events detected require RAG1 and its active site and are independent of RAG2.
As RAG1 and RAG2 are present in all jawed vertebrates, it was possible that species that branched off at different times during gnathostome evolution could have retained a RAG1 protein that was less dependent on RAG2 than mouse RAG1. However, RAG1 from five different vertebrates showed levels of activity similar to mouse RAG1, suggesting that dependence on RAG2 was acquired as an early event in the evolution of the jawed vertebrate immune system (Supplemental Fig. S1B).
The events observed in the absence of RAG2 in fibroblasts were recapitulated in the more physiological context of Abelson kinase transformed pre-B-cell lines (v-abl) with a similar recombination substrate (Fig. 1D) by combining the v-abl kinase inhibitor STI-571, which induces RAG expression, with constitutive retroviral expression of RAG1 in RAG2−/− cells and RAG2 in RAG1−/− cells (Supplemental Fig. S1C). Treatment with STI-571 also leads to recombination of the endogenous Igκ locus in v-abl cells, with Vκ–Jκ recombination being readily detectable in STI-571-treated RAG2−/− cells constitutively expressing retroviral RAG1 (Fig. 1E) using a PCR assay that detects recombination between roughly a third of the Vκ segments (Schlissel and Baltimore 1989) and either Jκ1 or Jκ2. The joints generated by RAG1 alone in v-abl cells were generally similar in structure to those generated by RAG1 and RAG2 together (Supplemental Table S1C–E; Supplemental Fig. S2C–F), although some differences were noted in the frequency of insertions at the joints (Supplemental Fig. S2D). Furthermore, the distribution of Vκ gene segments used in the recombination events mediated by RAG1 alone was similar to that used in RAG1/RAG2 recombination events (Supplemental Fig. S2G). Hence, RAG1 is capable of mediating recombination of an integrated substrate and an endogenous antigen receptor locus in the absence of RAG2.
Breakdown of the 12/23 rule in the absence of RAG2
The recombination activity of RAG1 in the fibroblast and lymphocyte lines supports the hypothesis that RAG1, at some point in its evolutionary history, was capable of functioning on its own. Our findings also provided an opportunity to gain insight into the evolutionary history of the RSS by testing whether RAG1's preferred substrate is a 12/23RSS pair, as is the case for RAG1 and RAG2.
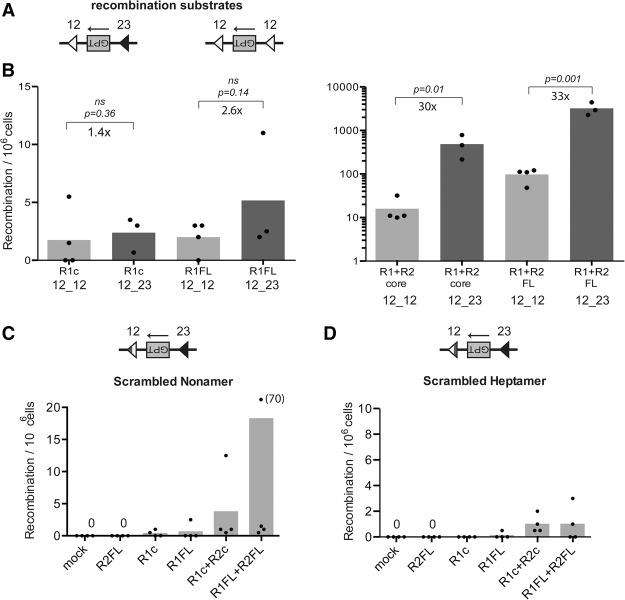
To test this, inversional substrates containing either a pair of 12RSSs or a 12RSS and a 23RSS (Fig. 2A) were introduced into RAG2−/− 3T3 fibroblasts. As expected, coexpressed RAG1 and RAG2 core or full length showed high levels of activity with the 12/23RSS substrate, with hundreds to thousands of events detected per million cells screened, while the activity dropped dramatically with the 12/12RSS substrate (∼15 and 100 events per million cells plated for core and full-length RAGs, respectively) (Fig. 2B, right panel). This represents a 30-fold and 33-fold change in recombination activity for core and full-length RAG, respectively. In contrast, RAG1 alone did not exhibit a strong preference for the 12/23 substrate. An average of about three and five events per million cells were observed for core and full-length RAG1, respectively, with the 12/23 substrate, and these numbers dropped only modestly to about two events per million with the 12/12 substrate (Fig. 2B, left panel). This yields a fold change in activity of 1.4 and 2.6 for core and full-length RAG1, respectively—differences that are not statistically significant. Several lines containing a 23/23RSS substrate were also tested, but all exhibited such low levels of activity with coexpressed RAG1 and RAG2 that they could not be used to gather reliable data with RAG1 alone (data not shown).
Figure 2.
RAG1 does not obey the 12/23 rule. (A) Simplified schematic of recombination substrates. The GPT gene is flanked by a pair of 12RSSs (white; 12_12 substrate) or one 12RSS and one 23RSS (black; 12_23 substrate). (B–D) Quantitation of recombination in RAG2−/− 3T3 fibroblast lines. Recombination efficiency is plotted as in Figure 1B. (B) Note the Y-axis scaling. (Left) Linear. (Right) Logarithmic. (C,D) Note the different Y-axis scales for the two plots and that, in C, one data point with a value of 70 was outside the range of the Y-axis, as indicated. (c) Core RAG proteins; (FL) full-length RAG proteins. One tailed t-test; P-values are noted. (ns) Not significant.
Given the less strict spacer length requirements for RAG1 activity, it was important to test whether the conserved elements of the RSS—the heptamer and the nonamer—were required for RAG1-mediated recombination. The heptamer specifies the site of cleavage, while the nonamer plays a significant role in RAG binding to the RSS, and mutations in the most conserved residues of either element greatly decrease recombination activity (Hesse et al. 1989). Wild-type 3T3 lines with substrates containing an intact 23RSS and a 12RSS with either a scrambled heptamer or nonamer were generated and revealed, as expected, poor activity using coexpressed RAG1 and RAG2, with only a few events observed per million cells screened (Fig. 2C,D). RAG1 alone showed virtually no activity with either substrate, highlighting the importance of both the heptamer and nonamer for the RAG2-independent reaction. Together, our data suggest an evolutionary role for RAG2 in helping to establish the 12/23 rule.
Transib and purple sea urchin RAG1-like can mediate V(D)J recombination with the help of RAG2
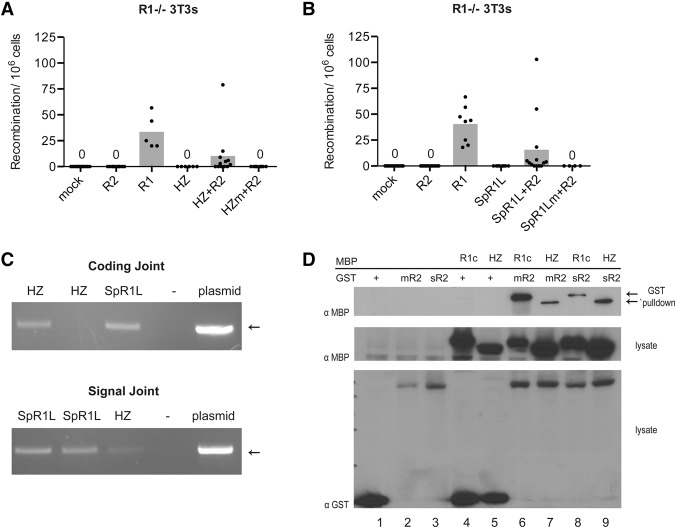
The ability of RAG1 to mediate recombination in the absence of RAG2 suggests the possibility that RAG1 was once an enzymatically independent protein, and several candidate TEs that have a sole RAG1-like transposase gene have been identified. Conversely, a number of RAG1-like genes with partner RAG2-like genes have also been identified in species that predate jawed vertebrates, although none have been demonstrated to have catalytic activity. The transition of a putative RAG1 predecessor from transposase to recombinase and the arrival of RAG2 remain little understood. To address these issues, two candidate proteins related to RAG1 were analyzed for V(D)J recombination activity. One, the protein encoded by the Transib from Helicoverpa zea (Hztransib), has been characterized as an active transposase with mechanistic similarities to RAG (Hencken et al. 2012). The other, a RAG1-like protein from Strongylocentrotus purpuratus (spRAG1L), is accompanied by a partner RAG2-like protein, but no recombination or transposition activity has yet been attributed to them (Fugmann et al. 2006). RAG1−/− 3T3 cells containing the 12/23RSS inversional substrate were transduced to express the candidate proteins or controls. When either Hztransib or spRAG1L was expressed in the fibroblasts, no recombination activity was detected. Strikingly, however, when either protein was coexpressed with mouse full-length RAG2, an average of 10 and 15 events per million cells screened were detected for the Transib and the RAG1-like protein, respectively—a level of activity approximately threefold lower than that observed for full-length RAG1 (Fig. 3A,B). Importantly, mutation of predicted active site residues in either protein abolished activity, demonstrating that the recombination events detected were the result of the action of the Hztransib and spRAG1L proteins (Fig. 3A,B). The integrity of the coding and signal joints was verified by PCR (Fig. 3C), and sequencing revealed coding and signal joints similar to those observed for coexpressed RAG1 and RAG2 (Supplemental Table S1F). Coexpression of spRAG1L and spRAG2L did not yield any recombination activity (data not shown). These results demonstrate that mouse RAG2 uncovers a latent ability in Hztransib and spRAG1L to cleave two RSSs precisely at their heptamer borders and thereby trigger inversional V(D)J recombination, leading to normal coding and signal joints.
Figure 3.
Recombination mediated by HzTransib and purple sea urchin RAG1-like. (A,B) Quantitation of recombination in RAG1−/− 3T3 fibroblast lines. Recombination efficiency is plotted as in Figure 1B. Note that levels of recombination by RAG1 alone in this RAG1−/− line were higher than in wild-type and RAG2−/− lines. (Mock) Empty vector control; (HZ) Hztransib; (SpR1L) spRAG1L; (HZm) active site mutant of Hztransib; (SpR1Lm) active site mutants of spRAG1L. (C) Representative PCR assays for coding or signal joints in genomic DNA from MPA-resistant cells expressing the indicated proteins. Each lane is an independent clone. (Plasmid) Reaction spiked with positive control template DNA; (HZ) Hztransib; (SpR1L) spRAG1L. (D) GST pull-down to detect interactions between the indicated MBP- or GST-tagged proteins. (R1c) RAG1 core; (Hz) HZTransib; (mR2) mouse RAG2; (sR2) shark RAG2; (+) GST alone. (Top) Glutathione pull-down. (Middle and bottom) Starting material lysate. Arrows indicate the expected positions of MBP-RAG1 core and MBP-Hztransib proteins.
To begin to understand how RAG2 activates the RSS cleavage activity of these enzymes, we tested for a physical interaction between Hztransib and RAG2. MBP-tagged Hztransib was coexpressed with GST-tagged mouse or shark RAG2 followed by pull-down with a GST affinity resin and blotting with anti-MBP antibodies. MBP-tagged mouse RAG1 core and GST served as positive and negative controls, respectively. As expected, MBP-RAG1 core interacted well with mouse RAG2 and detectably with shark RAG2 (Fig. 3D, lanes 6,8). An interaction was also readily detected between Hztransib and both RAG2 proteins (Fig. 3D, lanes 7,9). These results, together with previous findings showing an interaction between spRAG1L and shark RAG2 (Fugmann et al. 2006), are consistent with a model in which the capacity of an ancient RAG1-like protein to interact with RAG2 enhanced its ability to perform recombination and highlight the potential importance of RAG2 for the transition of RAG1 from transposase to recombinase.
RAG2 enhances transposition by Hztransib
The RAG2–Hztransib interaction suggested that RAG2 might directly stimulate the RSS-mediated recombination activity of Hztransib by altering its biochemical properties. This in turn suggested that RAG2 might influence the ability of Hztransib to integrate a TIR-flanked donor DNA fragment into host DNA. To test this, we used an in vitro transposition assay (Agrawal et al. 1998) involving incubation of purified proteins with a target plasmid and a donor DNA fragment terminating in 5′ phosphorylated TIRs or RSSs that mimics a cleaved substrate prior to the strand transfer step of transposition (Fig. 4A). Insertion of the donor fragment harboring TetR into the target plasmid containing KanR generates a plasmid easily identified after transformation into bacteria by selection on tetracycline and kanamycin.
Figure 4.
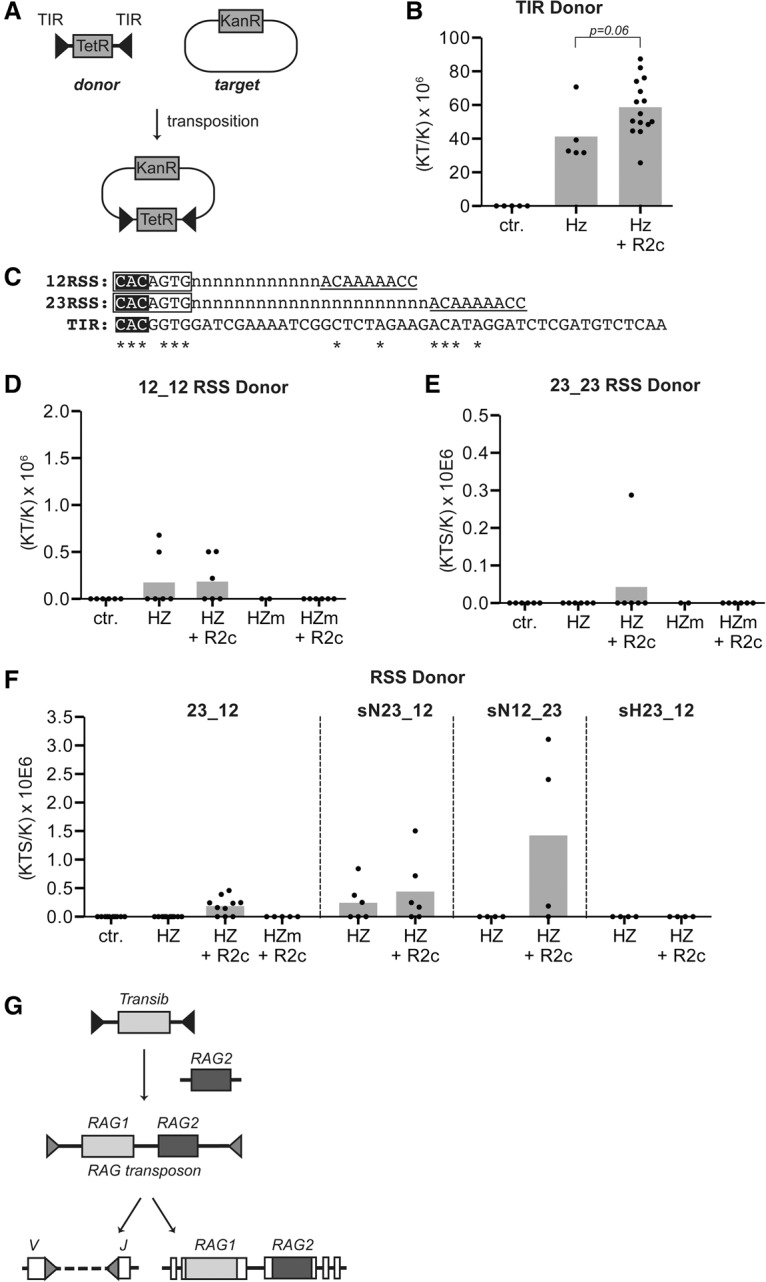
Transib transposition is enhanced by RAG2. (A) Schematic of in vitro transposition assay. Transposition of a donor DNA fragment, a tetracycline resistance gene (TetR) flanked by pair of TIRs or RSSs (black triangles), into target plasmid with the kanamycin resistance gene (KanR) generates double resistance plasmids. (B,D–F) Quantitation of transposition activity as measured after bacterial transformation. The transposition efficiency for the indicated protein or proteins is plotted as the number of transposition events (Kan/Tet double-resistant colonies) normalized to the total amount of plasmid purified from each reaction (Kan-resistant colonies) and multiplied by 106, with each dot representing an independent experiment and the bar height indicating the mean. (Ctr.) No protein; (HZ) Hztransib; (HZm) Hztransib active site mutant; (R2c) RAG2 core. (B) TIR donor; two-tailed t-test. (D,E) RSS donors. (sN) Scrambled nonamer; (sH) scrambled heptamer. In mutant RSS substrates, the heptamer or nonamer was scrambled in one of the two RSSs, with the other RSS remaining intact. (C) Alignment of Hztransib TIRs to consensus 12RSS and 23RSS. (Boxed) Heptamer; (underlined) nonamer; (highlighted) conserved positions of the heptamer crucial for activity; (*) positions conserved between the RSS and Transib TIR. (G) Model for the origin of the RAG proteins and V(D)J recombination. We propose a two-step model in which a Transib element acquired an ancestral RAG2 gene, leading to a “RAG transposon” containing both RAG1 and RAG2, whose disassembled components subsequently gave rise to split antigen receptor genes and the RAG locus of jawed vertebrates, as proposed by Thompson (1995).
As expected (Hencken et al. 2012), Hztransib was capable of transposing a donor fragment flanked by TIRs (Fig. 4B), and, consistent with prior findings (Kapitonov and Jurka 2005; Hencken et al. 2012), the target site duplications (TSDs) were invariably 5 base pairs in length (Supplemental Table S1G) and slightly enriched for GC base pairs (Supplemental Fig. S3D). Interestingly, coincubation with RAG2 core yielded a strong trend toward increased transposition activity by Hztransib (Fig. 4B), with TSDs similar to those generated without RAG2 (Supplemental Fig. S3A).
Hztransib TIRs have clear sequence similarity to the RSS heptamer but little or none to the RSS nonamer (Fig. 4C). Given this and the recombination activity observed on RSSs, Hztransib was tested for its ability to transpose an RSS substrate. When a substrate containing a 12/12RSS pair was used, transposition by Hztransib was observed in the absence or presence of RAG2 (Fig. 4D), but activity was difficult to detect under either condition using a 23/23RSS substrate (Fig. 4E). Notably, when a 12/23RSS pair was used, transposition activity was observed only in the presence of RAG2 (Fig. 4F). These data suggest that Hztransib has more difficulty performing transposition with a 23RSS than a 12RSS. Data consistent with this idea were obtained using substrates in which the nonamer of one RSS was scrambled: When the 12RSS was left intact, Hztransib was active in the presence and absence of RAG2 (Fig. 4F, substrate sN23_12), but when the 23RSS was left intact, Hztransib was active only when coincubated with RAG2 (Fig. 4F, substrate sN12_23). Together, the data suggest that RAG2 assists Hztransib to perform transposition of substrates containing a 23RSS and that Hztransib is sensitive to the presence and location of the nonamer despite lacking most of the region that corresponds to the vertebrate RAG1 nonamer-binding domain (Supplemental Fig. S3F). Consistent with the critical role played by the invariant 5′ CAC at the beginning of the heptamer for RAG activity, Hztransib transposase activity was eliminated by scrambling the heptamer of one RSS (Fig. 4F). Also consistent with the importance of the heptamer, a greater proportion of heptamer contact residues in RAG1 (Ru et al. 2015) are identical in Hztransib than residues overall (44% vs. 22%) (Supplemental Fig. S3F). Characterization of the target sites used by Hztransib with RSS substrates revealed a tightly restricted distribution at sites composed almost entirely of G and C (Supplemental Table S1G; Supplemental Fig. S3B,C,E). Therefore, similar to the role observed for RAG2 in helping establish the 12/23 rule for V(D)J recombination, it appears that RAG2 plays a related role in helping Hztransib carry out the strand transfer step of transposition with substrates containing a 23RSS. Hence, it is plausible that RAG2 reinforced the preference for asymmetrical substrates by helping the ancestral Transib element accommodate the 23RSS.
Discussion
Based on our findings, we propose a model for the evolution of V(D)J recombination in which a Transib transposase gene was the evolutionary precursor of RAG1, and acquisition of an ancestral RAG2 by a Transib element gave rise to the RAG1–RAG2 transposon (Fig. 4G). The origins of RAG2 remain unclear, with no candidate RAG2 precursor having yet been identified independent of a flanking RAG1-like gene. It is plausible, however, that the ancestral RAG2 was a host factor already encoded in the genome of the organism in which the RAG transposon first arose (perhaps an early deuterostome). We propose that acquisition of RAG2 provided functional advantages to the ancestral Transib while also increasing the range of activities available to it. Close partnership with RAG2 not only might have enhanced the transposition activity of Transib, but the novel capacity of the RAG1–RAG2 gene pair to mediate recombination could have allowed for its positive selection and evolution into the recombinase required to assemble a split antigen receptor gene.
In addition, RAG2 might have played a role in establishing the 12/23 rule, first by helping the ancient Transib accommodate a 23RSS for transposition and later in helping maintain the preference for an asymmetrical pair of RSSs during the recombination reaction. As the 12/23 rule imposes order on the V(D)J recombination process and helps ensure the inclusion of certain gene segments, its benefits could have been selected for during the evolution of this system. The contribution of RAG2 to establishing this rule would have provided yet another selective pressure for maintaining this second protein in the ancient recombinase complex. The mechanism by which RAG2 facilitates the 12/23 rule is not known, but a recent structural study (Ru et al. 2015) provided potentially relevant insights, most notably that RSS binding induces conformational changes in the RAG complex. These changes include new RAG1–RAG2 interactions important for the formation of a “closed” RAG–RSS synaptic complex, presumed to be the fully active conformation. The stabilization imparted by RAG2 to the complex and the additional DNA contacts that it provides might have played a role in limiting the ability of the RAG complex to accommodate a symmetrical pair of RSSs.
It was extremely difficult to detect V(D)J recombination in STI-571-induced RAG2−/− v-abl pre-B-cell lines unless exogenous RAG1 was also provided from a retroviral vector (Fig. 1E; data not shown). Because the increase in RAG1 expression attributable to the retrovirus was more apparent prior to than after induction (Supplemental Fig. S1C), we cannot determine whether the strong enhancement in recombination linked to retrovirally derived RAG1 was due to post-induction overexpression of RAG1 or the higher expression levels in the uninduced cells. Our results and those from many studies of RAG2−/− mice (e.g., Shinkai et al. 1992) argue that RAG1 levels are sufficiently low in developing lymphocytes that RAG1-only recombination is exceedingly rare. A recent study estimated that mouse thymocytes contain, on average, only ∼1000 RAG1 dimers (Zhang et al. 2015).
It is possible that recombination mediated by RAG1 in the absence of RAG2 is only detectable with a powerful selection, as was the case in our experiments and in a study involving transfer of RAG2-deficient T cells into allogeneic RAG2−/− hosts, which identified transferred cells that no longer expressed the expected transgenic T-cell receptor (TCR) but instead expressed a different TCR specificity (Montaudouin et al. 2010). The low frequency of recombination in the absence of RAG2 along with the strong stimulatory effect of ectopic RAG1 expression helps reconcile our findings with the lack of reports of recombination events in developing lymphocytes from unmanipulated RAG2−/− mice. Nevertheless, we cannot exclude the possibility that “RAG1-only” recombination in cultured cells and mice is due to a host factor that weakly compensates for the absence of RAG2.
The ability of RAG1 to generate coding and signal joints similar in most respects to those generated in the presence of RAG2 has several implications. First, the presence of joints with intact coding and signal ends reveals the ability of RAG1 to cut DNA precisely at the border between the RSS heptamer and the coding segment DNA, the same location that is cut in the presence of RAG2. In this regard, it is notable that the recently reported structure of RAG bound to DNA reveals that all direct contacts with the RSS are made by RAG1, while RAG2 makes a small number of contacts with flanking coding segment DNA (Ru et al. 2015). Second, it can be inferred from the presence of P nucleotides in the coding joints generated by RAG1 that the cleavage reaction proceeds through a nick-hairpin mechanism, as has been documented for RAG1 with RAG2. Third, the relatively normal structure of coding and signal joints implies that, in the absence of RAG2, the cut DNA ends can be repaired by the nonhomologous end-joining machinery, albeit with increased activity of TdT at coding and signal ends. Finally, detection of recombination strongly implies that RAG1 is capable of making breaks at the two RSSs sufficiently close in time to allow for the formation of recombinants. Thus far, it has not been possible to detect RAG1-mediated DNA cleavage in vitro, which would be necessary for a more detailed mechanistic analysis of DNA cleavage in the absence of RAG2.
The evolutionary timing of the appearance of the RAG transposon remains uncertain. Our discovery that spRAG1L, a putative RAG1 relative in echinoderms, possesses catalytic activity with functional parallels to RAG1 from jawed vertebrates provides important support for the hypothesis that a RAG TE was present in the common ancestor of deuterostomes (Fugmann et al. 2006; Fugmann 2010), an idea that received additional support from a recent analysis of several new echinoderm genome assemblies (Kapitonov and Koonin 2015). Such a model implies vertical transfer of a RAG1/RAG2 gene pair in deuterostomes with its loss from lineages in which no RAG-like genes can be found (e.g., agnathans) (Hirano et al. 2011), although the occurrence of several independent horizontal transfer events cannot be ruled out. By providing clear functional evidence for a relationship between RAG and its proposed transposase predecessors, our results argue against convergent evolution of vertebrate RAG1 and purple sea urchin RAG1-like and support a two step model for the evolution of the RAG recombinase (Fig. 4G).
Materials and methods
DNA substrates and inversional recombination assay
Retroviral recombination substrates were integrated into 3T3 or v-abl lines (for details, see the Supplemental Material), and cells were selected and subcloned to obtain clonal populations for subsequent transductions with protein expression vectors. After selection for cells containing protein expression vectors, 3T3 cells were plated in MPA-containing medium as previously described (Schatz and Baltimore 1988). After 12 d in selection, MPA-resistant clones were expanded for further analysis and/or counted after Crystal Violet staining. The 12/12 and 12/23 retroviral recombination substrates were constructed as described in the Supplemental Material.
PCR assays for coding and signal joints
For 3T3 lines, genomic DNA was harvested from individual clones or bulk populations of MPA-resistant cells. v-abl cell genomic DNA was harvested after 72 h of incubation with STI-571. Either a single round of PCR (signal joints from the ZGR substrate) or nested PCR (all other joints) was performed using the primers and amplification conditions described in the Supplemental Material and Supplemental Table S1H. PCR products were gel-extracted or TOPO-TA-cloned for sequencing.
GST pull-downs and immunoblotting
RAG1, RAG2, and Transib pEBB expression vectors were transfected into 293T cells using calcium phosphate. Lysates were prepared, and pull-downs were performed as previously described (Zhang et al. 2015). Immunoblots were developed with anti-GST (Cell Signaling) and anti-MBP (New England Biolabs) antibodies.
Protein purification
RAG1 core, RAG2 core, and Hztransib were cloned into pTT5 (Durocher et al. 2002), which provides an N-terminal MBP fusion tag, and were expressed in expi293 cells (ThermoFisher) using the company's protocol. All proteins were purified on amylose affinity columns and dialyzed in storage buffer (20 mM Tris at pH 8, 150 mM KCl, 2 mM DTT). HMGB1 was purified as previously described (Eastman et al. 1999).
In vitro transposition assay
Transposition reactions were performed essentially as previously described (Agrawal et al. 1998). For modifications, see the Supplemental Material. Donor fragments were generated with 5′ phosphorylated primers.
Supplementary Material
Acknowledgments
We thank Nancy Craig for providing the Hztransib ORF, Vladimir Kapitonov for useful discussions regarding Transib; Shaoming Zhou for help with maintenance of cells lines, Tony Koleske for guidance on the preparation of 3T3 lines, and Yuhang Zhang for help with protein purification and helpful discussion. This work was supported in part by a National Institutes of Health National Research Service Award Institutional Predoctoral Training grant (T32 AI007019) and the Yale Immunobiology Department Gershon Fellowship. S.D.F. was supported by a Chang Gung Memorial Hospital grant (CMRPD1C0242). D.G.S. is an investigator of the Howard Hughes Medical Institute. L.M.C. and D.G.S. designed the experiments. L.M.C. performed the experiments. L.M.C., D.G.S., and S.D.F. analyzed and discussed the data. L.M.C. and D.G.S. wrote the paper.
Footnotes
Supplemental material is available for this article.
Article published online ahead of print. Article and publication date are online at http://www.genesdev.org/cgi/doi/10.1101/gad.278432.116.
References
- Agrawal A, Eastman QM, Schatz DG. 1998. Transposition mediated by RAG1 and RAG2 and its implications for the evolution of the immune system. Nature 394: 744–751. [DOI] [PubMed] [Google Scholar]
- Durocher Y, Perret S, Kamen A. 2002. High-level and high-throughput recombinant protein production by transient transfection of suspension-growing human 293-EBNA1 cells. Nucleic Acids Res 30: E9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Eastman QM, Villey IJ, Schatz DG. 1999. Detection of RAG protein–V(D)J recombination signal interactions near the site of DNA cleavage by UV cross-linking. Mol Cell Biol 19: 3788–3797. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fugmann SD. 2010. The origins of the Rag genes—from transposition to V(D)J recombination. Semin Immunol 22: 10–16. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fugmann SD, Messier C, Novack LA, Cameron RA, Rast JP. 2006. An ancient evolutionary origin of the Rag1/2 gene locus. Proc Natl Acad Sci 103: 3728–3733. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hencken CG, Li X, Craig NL. 2012. Functional characterization of an active Rag-like transposase. Nat Struct Mol Biol 19: 834–836. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hesse JE, Lieber MR, Mizuuchi K, Gellert M. 1989. V(D)J recombination: a functional definition of the joining signals. Genes Dev 3: 1053–1061. [DOI] [PubMed] [Google Scholar]
- Hiom K, Melek M, Gellert M. 1998. DNA transposition by the RAG1 and RAG2 proteins: a possible source of oncogenic translocations. Cell 94: 463–470. [DOI] [PubMed] [Google Scholar]
- Hirano M, Das S, Guo P, Cooper MD. 2011. The evolution of adaptive immunity in vertebrates. Adv Immunol 109: 125–157. [DOI] [PubMed] [Google Scholar]
- Ji Y, Resch W, Corbett E, Yamane A, Casellas R, Schatz DG. 2010. The in vivo pattern of binding of RAG1 and RAG2 to antigen receptor loci. Cell 141: 419–431. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jones JM, Gellert M. 2004. The taming of a transposon: V(D)J recombination and the immune system. Immunol Rev 200: 233–248. [DOI] [PubMed] [Google Scholar]
- Kapitonov VV, Jurka J. 2005. RAG1 core and V(D)J recombination signal sequences were derived from Transib transposons. PLoS Biol 3: e181. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kapitonov VV, Koonin EV. 2015. Evolution of the RAG1–RAG2 locus: both proteins came from the same transposon. Biol Direct 10: 20. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kim MS, Lapkouski M, Yang W, Gellert M. 2015. Crystal structure of the V(D)J recombinase RAG1–RAG2. Nature 518: 507–511. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Liu Y, Subrahmanyam R, Chakraborty T, Sen R, Desiderio S. 2007. A plant homeodomain in RAG-2 that binds Hypermethylated lysine 4 of histone H3 is necessary for efficient antigen-receptor-gene rearrangement. Immunity 27: 561–571. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Matthews AG, Kuo AJ, Ramon-Maiques S, Han S, Champagne KS, Ivanov D, Gallardo M, Carney D, Cheung P, Ciccone DN, et al. 2007. RAG2 PHD finger couples histone H3 lysine 4 trimethylation with V(D)J recombination. Nature 450: 1106–1110. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mombaerts P, Iacomini J, Johnson RS, Herrup K, Tonegawa S, Papaioannou VE. 1992. RAG-1-deficient mice have no mature B and T lymphocytes. Cell 68: 869–877. [DOI] [PubMed] [Google Scholar]
- Montaudouin C, Boucontet L, Mailhe-Lembezat MP, Mariotti-Ferrandiz ME, Louise A, Six A, Freitas AA, Garcia S. 2010. Endogenous TCR recombination in TCR Tg single RAG-deficient mice uncovered by robust in vivo T cell activation and selection. PLoS One 5: e10238. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Notarangelo LD, Villa A, Schwarz K. 1999. RAG and RAG defects. Curr Opin Immunol 11: 435–442. [DOI] [PubMed] [Google Scholar]
- Oettinger MA, Schatz DG, Gorka C, Baltimore D. 1990. RAG-1 and RAG-2, adjacent genes that synergistically activate V(D)J recombination. Science 248: 1517–1523. [DOI] [PubMed] [Google Scholar]
- Ramsden DA, Baetz K, Wu GE. 1994. Conservation of sequence in recombination signal sequence spacers. Nucleic Acids Res 22: 1785–1796. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rooney S, Chaudhuri J, Alt FW. 2004. The role of the non-homologous end-joining pathway in lymphocyte development. Immunol Rev 200: 115–131. [DOI] [PubMed] [Google Scholar]
- Ru H, Chambers MG, Fu TM, Tong AB, Liao M, Wu H. 2015. Molecular mechanism of V(D)J recombination from synaptic RAG1–RAG2 complex structures. Cell 163: 1138–1152. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schatz DG, Baltimore D. 1988. Stable expression of immunoglobulin gene V(D)J recombinase activity by gene transfer into 3T3 fibroblasts. Cell 53: 107–115. [DOI] [PubMed] [Google Scholar]
- Schatz DG, Swanson PC. 2011. V(D)J recombination: mechanisms of initiation. Annu Rev Genet 45: 167–202. [DOI] [PubMed] [Google Scholar]
- Schatz DG, Oettinger MA, Baltimore D. 1989. The V(D)J recombination activating gene, RAG-1. Cell 59: 1035–1048. [DOI] [PubMed] [Google Scholar]
- Schlissel MS, Baltimore D. 1989. Activation of immunoglobulin κ gene rearrangement correlates with induction of germline κ gene transcription. Cell 58: 1001–1007. [DOI] [PubMed] [Google Scholar]
- Shinkai Y, Rathbun G, Lam KP, Oltz EM, Stewart V, Mendelsohn M, Charron J, Datta M, Young F, Stall AM, et al. 1992. RAG-2-deficient mice lack mature lymphocytes owing to inability to initiate V(D)J rearrangement. Cell 68: 855–867. [DOI] [PubMed] [Google Scholar]
- Swanson PC. 2004. The bounty of RAGs: recombination signal complexes and reaction outcomes. Immunol Rev 200: 90–114. [DOI] [PubMed] [Google Scholar]
- Teng G, Maman Y, Resch W, Kim M, Yamane A, Qian J, Kieffer-Kwon KR, Mandal M, Ji Y, Meffre E, et al. 2015. RAG represents a widespread threat to the lymphocyte genome. Cell 162: 751–765. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Thompson CB. 1995. New insights into V(D)J recombination and its role in the evolution of the immune system. Immunity 3: 531–539. [DOI] [PubMed] [Google Scholar]
- Zhang Y, Xu K, Deng A, Fu X, Xu A, Liu X. 2014. An amphioxus RAG1-like DNA fragment encodes a functional central domain of vertebrate core RAG1. Proc Natl Acad Sci 111: 397–402. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhang YH, Shetty K, Surleac MD, Petrescu AJ, Schatz DG. 2015. Mapping and quantitation of the interaction between the recombination activating gene proteins RAG1 and RAG2. J Biol Chem 290: 11802–11817. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.