Figure 2.
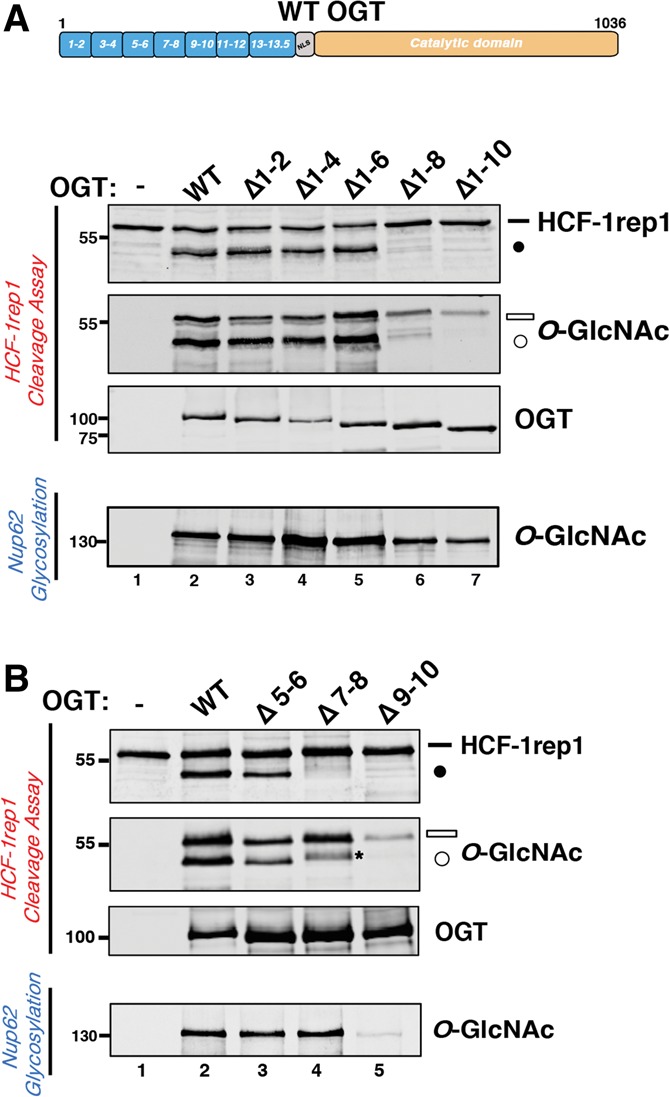
TPR domain requirements for HCF-1 proteolysis. (A) Mutant OGTs containing progressive N-terminal TPR deletions (schematic representation shown in Supplemental Fig. 2A) were incubated with HCF-1rep1 or Nup62 and assayed for in vitro HCF-1rep1 cleavage and glycosylation or Nup62 glycosylation as described in the Materials and Methods. HCF-1rep1 cleavage was detected with anti-GST antibody, whereas anti-O-GlcNAc RL2 was used to detect HCF-1rep1 and Nup62 glycosylation. Anti-T7 tag antibody was used to detect OGT. (B) Mutant OGTs containing internal deletions of two TPR motifs at a time (schematic representation shown in Supplemental Fig. 2B) were incubated with HCF-1rep1 or Nup62 and assayed for in vitro HCF-1rep1 cleavage and glycosylation or Nup62 glycosylation as described in A. The asterisk identifies a contaminating background band described in the Supplemental Material. (Black bar) Uncleaved HCF-1rep1 substrate; (black circle) cleaved product; (white bar) glycosylated uncleaved HCF-1rep1 substrate; (white circle) glycosylated cleaved product.
