Abstract
The aim of the present study was to investigate the association between microRNA-141 (miR141) and signal transducer and activator of transcription 5 (STAT5) expression levels in human esophageal squamous cell carcinoma (ESCC) and to investigate the effects of miR141 on ESCC cells. A total of 45 consecutive patients with ESCC were enrolled in the study. The expression of miR141 in ESCC tissue samples was detected using reverse transcription quantitative polymerase chain reaction (RT-qPCR). The expression of STAT5 in the ESCC tissues was detected using immunohistochemical staining and western blotting. In addition, Eca109 cells were transfected with miR141 mimic, and the levels of STAT5 were detected using western blotting. The effects of miR141 on the proliferation, invasion and migration of the cells were also detected using MTT, scratch and Transwell invasion assays, respectively. The miR141 expression level in the ESCC tissue samples was significantly decreased compared with that in the adjacent normal tissues (P<0.05). The expression of miR141 in the tissues from patients with lymph node metastasis was significantly decreased compared with that in the tissues of patients without such metastasis (P<0.05). The expression levels of STAT were significantly increased in the ESCC tissues compared with those in the adjacent normal tissues (P<0.05). Furthermore, the levels of STAT5 were significantly increased in the tissues from patients with lymph node metastasis compared with those without such metastasis (P<0.05); however, no statistically significant differences in miR141 expression were observed according to gender, age, tumor size, lesion location, differentiation and invasion (P>0.05). The results suggest that the miR141 mimic significantly inhibited the proliferation, migration and invasion of Eca109 cells in vitro. miR141 and STAT5 expression levels exhibited a negative association in the ESCC tissues, and were both closely associated with the progression of ESCC. Therefore, it appears that miR141 plays an important role in the development, invasion and metastasis of ESCC by regulating the expression of STAT5.
Keywords: esophageal squamous cell carcinoma, miR141, signal transducer and activator of transcription 5, invasion, metastasis
Introduction
Esophageal squamous cell carcinoma (ESCC) is an upper gastrointestinal malignancy, characterized by insidious onset, rapid development and poor prognosis (1). Since the rate of early diagnosis is poor, the majority of patients are diagnosed while at the advanced stage of ESCC, and their 5-year survival rates are 10–30%. Invasion and metastasis of ESCC are the main reasons for poor prognosis following surgery (2). Early diagnosis, and anti-invasion and anti-metastasis therapies can significantly improve the prognosis of ESCC. Therefore, it is necessary to investigate the pathogenic mechanisms of ESCC.
microRNA (miRNA) is endogenous small non-coding RNA that modulates gene expression. The abnormal expression of microRNA plays an important role in tumorigenesis (3). The level of a particular miRNA, miR141, has been found to be upregulated in breast, lung and stomach tumors and downregulated in liver and prostate tumors (4–6). miR141 is known to be involved in the proliferation, invasion, apoptosis and angiogenesis of gastric, liver and pancreatic tumors (7). In the present study, the target gene of miR141 was predicted to be signal transducer and activator of transcription 5 (STAT5) using bioinformatic analysis.
STAT5, an important signal transducer and transcription activator, has an important role in the Janus kinase/STAT signaling pathway (8). Its expression level has been shown to be significantly increased in tumors, where it is directly involved in differentiation, invasion, proliferation and apoptosis (9–11).
In the present study, the expression of miR141 in clinical samples of ESCC was detected using reverse transcription quantitative polymerase chain reaction (RT-qPCR). The levels of STAT5 were detected in the ESCC tissues by immunohistochemical staining and western blotting. The effects of miR141 on the proliferation, migration and invasion of Eca109 human esophageal epithelial cancer cells were detected with the use of 3-(4,5-dimethyl-2-thiazolyl)-2,5-diphenyl-2H-tetrazolium bromide (MTT), scratch and Transwell invasion assays, respectively.
Materials and methods
Clinical data of patients
A total of consecutive 45 patients (37 male and 8 female; mean age, 58.30±8.10 years; age range, 42–71 years) pathologically diagnosed with ESCC were enrolled in the study. Among these 45 patients, 28 cases had well-differentiated tumors, 14 cases had moderately differentiated tumors and 3 cases had poorly differentiated tumors. Lymph node metastasis was identified in 21 cases and the other 24 cases were not found to have lymph node metastasis. All patients underwent surgical treatment. Esophageal cancer tissues and peritumoral tissues (≥5 cm away from the tumor edge) were collected during surgery. Tumor size, differentiation and invasion were evaluated by pathologists. Lesion location was identified by endoscopic examination and during surgery. All tissues were frozen in liquid nitrogen and then stored at −80°C until further analysis. Information about the patients is shown in Table I. None of the patients had a history of chemotherapy or additional tumors. Prior written and informed consent was obtained from every patient and the study was approved by the ethics review board of Linyi People's Hospital (Linyi, China).
Table I.
Classification of the patients with esophageal squamous cell carcinoma (n=45).
| Classification | No. of cases |
|---|---|
| Degree of differentiation | |
| High differentiation | 28 |
| Moderate differentiation | 14 |
| Poor differentiation | 3 |
| Metastatic status | |
| Lymph node metastasis | 21 |
| Without lymph node metastasis | 24 |
Bioinformatic analysis
The target gene of miR141 was predicted using TargetScan (http://www.targetscan.org) and miRwalk (http://www.umm.uniheidelberg.de/apps/zmf/mirwalk) databases, and pathway analysis was conducted using Kyoto Encyclopedia of Genes and Genomes (KEGG; http://www.genome.jp/kegg/) and Gene Ontology (GO; (http://www.geneontology.org) databases.
Reagents
Reagents used included TRIzol reagent (Invitrogen; Thermo Fisher Scientific, Inc., Waltham, MA, USA), rabbit anti-human STAT5 polyclonal antibody (ab68465; Abcam, Cambridge, MA, USA), a streptavidin-peroxidase (SP) kit (Beijing Zhongshan Golden Bridge Biotechnology Co., Ltd., Beijing, China), PrimeScript™ RT reagent kit and SYBR® PrimeScript™ real-time PCR kit (both Takara Biotechnology Co., Ltd., Dalian, China).
Cell culture and miRNA transfection
Eca109 cells (Cell Bank of the Chinese Academy of Sciences, Shanghai, China) were cultured at a concentration of 2×105 cells/well in 24-well plates with RPMI-1640 (Hyclone; GE Healthcare Life Sciences, Logan, UT, USA) supplemented with 10% fetal bovine serum (Gibco FBS; Thermo Fisher Scientific, Inc.). When 70–80% confluence was reached, the cells were divided into the normal control, negative control (NC) and miR141 mimic groups. Cells in the NC group remained untransfected, while cells in the NC and miR141 groups were transfected with miR-NC and miR141, respectively, by lipofection. In brief, 1.5 µl miRNA mimic or miR-NC (both 20 pmol/µl; Guangzhou RiboBio Co., Ltd., Guangzhou, China), together with 1 µl Lipofectamine 2000 (Invitrogen; Thermo Fisher Scientific, Inc.) was added to an EP tube supplemented with 50 µl Opti-MEM medium (Invitrogen; Thermo Fisher Scientific, Inc.), and allowed to stand undisturbed for 5 min. The mixture was then added to the cells, and the cells were incubated for 6 h. The medium was then replaced with RPMI-1640 supplemented with 10% FBS for further culture. The protein expression level of STAT5 was detected at 48 h.
Immunohistochemical staining
Tumor tissues were fixed in 10% formaldehyde, embedded in paraffin and then cut into 4-µm sections. The sections were dewaxed in graded xylene, dehydrated in graded ethanol, and then incubated with 3% hydrogen peroxide for 10 min at room temperature. Antigen retrieval was conducted by incubation with sodium citrate (pH 6.0) at 95°C for 15 min. Following antigen retrieval, the sections were treated with anti-STAT5 antibody (l:200) for 1 h at room temperature, followed by incubation with biotinylated secondary anti-IgG (PK-4001; 1:200; Beijing Zhongshan Golden Bridge Biological Technology Co., Ltd.) for 30 min at 37°C. Subsequently, the sections were stained with 3,3′-diaminobenzidine, followed by slightly counterstaining with hematoxylin for 30 sec. Following differentiation with hydrochloric acid and treatment with xylene, sections sealed in neutral gum were observed under a CX31 microscope (Olympus Corporation, Tokyo, Japan).
Immunohistochemical grading
Brown staining of the cytoplasm or membrane was considered as STAT5-positive. Five fields of view were randomly selected. The percentage of positive cells was determined from the ratio of the number of positive cells relative to the total number of cells. At least 200 cells were counted.
RT-qPCR
Total RNA was extracted from the tissue samples using TRIzol reagent and reverse transcribed into cDNA using the PrimeScript™ RT reagent kit. Completeness of the cDNA was detected using gel electrophoresis and purity was detected using a spectrophotometer (NanoDrop Technologies, Inc., Wilmington, DE, USA) to measure the 260/280 absorbance ratio. The levels of miR141 and STAT5 were measured. The forward primer sequence for miR141 was 5′-CACTGTCTGGUAAAGA-3′. The reverse primer sequence for miR141 was universal. The primer sequences for STAT5 were as follow: Forward, 5′-ATTCAAACACTTGACCCTGA-3′ and reverse, 5′-TGTGTCCTCCAGATCGAAG-3′. Glyceraldehyde 3-phosphate dehydrogenase (GAPDH) was used as an internal control and the primer sequences were as follows: Forward, 5′-AGGTCGGTGTGAACGGATTTG-3′ and reverse, 5′-GGGGTCGTTGATGGCAACA-3′. PCR conditions were as follows: Denaturation for 10 min at 95°C, followed by 40 cycles of annealing for 1 min at 95°C and extension for 30 sec at 60°C. The dissolution procedure was designed as follows: 15 sec at 95°C, 30 sec at 60°C and 15 sec at 95°C. The PCR procedure was conducted using the SYBR® PrimeScript™ real-time PCR kit and a StepOnePlus™ Real-time PCR instrument (Applied Biosystems; Thermo Fisher Scientific, Inc.). The 2﹣ΔΔCq method was used to calculate the relative expression levels.
Western blot analysis
Proteins were extracted from tissues or cells by lysis using radioimmunoprecipitation assay (RIPA) buffer (Beyotime Institute of Biotechnology, Beijing, China). Sodium dodecyl sulfate-polyacrylamide gel electrophoresis (SDS-PAGE) sample buffer was added to the proteins, and the mixture was boiled for 5 min. Subsequently, 10 µl sample was separated using 10% SDS-PAGE gel (Beyotime Institute of Biotechnology) at 80 V, followed by electrophoretic transfer for 2 h at 200 mA to polyvinylidene difluoride membranes. The membranes were blocked with 50 g/l skimmed milk for 1 h at room temperature. Rabbit anti-human STAT5 antibody (1:1,000) or rabbit anti-GAPDH antibody (1:5,000; ab181602; Abcam) was added and the membrane was incubated overnight at 4°C. Then, goat anti-rabbit (1:2,000; ab6721; Abcam) secondary antibody was added and the membrane was incubated for 1 h at room temperature. Protein bands were visualized by electrochemiluminescence reaction (Beyotime Institute of Biotechnology). The results were quantified using Quantity One software, version 4.62 (Bio-Rad Laboratories, Inc., Hercules, CA, USA
MTT assay
Following transfection, Eca109 cells were cultured in 96-well plates at a density of 2×103/well for 0, 24, 48 and 72 h. Subsequently 20 µl 5 mg/l MTT was added and the cells were incubated for 4 h at 37°C. Finally, cell proliferation was calculated at an absorbance of 490 nm using an iMark absorbance reader (Bio-Rad Laboratories, Inc.).
Scratch assay
Eca109 cells (5×105) were cultured in 24-well plates for 24 h. A sterile pipette tip was used to draw straight lines along the longitudinal axis and then the cells were washed with PBS. The migration of the cells was examined under an IX83 inverted microscope (magnification, ×10; Olympus Corporation) at 24 h.
Transwell assay
After thawing overnight at 4°C, Matrigel (dilution, 1:2; BD Biosciences, Franklin Lakes, NJ, USA) was added to the upper chamber of a Transwell (Corning, Inc., Corning, NY, USA) and incubated for 60 min at 37°C. Eca109 cells (1×105) and 200 µl serum-free RPMI-1640 were added to the upper chamber and 500 µl RPMI-1640 supplemented with 10% FBS was added to the lower chamber and the Transwell unit was incubated for 24 h. Then, the cells that had crossed the membrane into the lower chamber were fixed with 4% formaldehyde for 10 min and stained with Giemsa. The extent of the cell invasion was determined by counting the transmembranous cells under a microscope.
Statistical analysis
All data were analyzed using SPSS software, version 11.0. (SPSS, Inc., Chicago, IL, USA). Results are presented as the mean ± standard deviation. Student's t-test was performed to compare differences between groups. P<0.05 was considered to indicate a statistically significant difference.
Results
Expression of miR141 is decreased in ESCC and in cases with lymph node metastasis
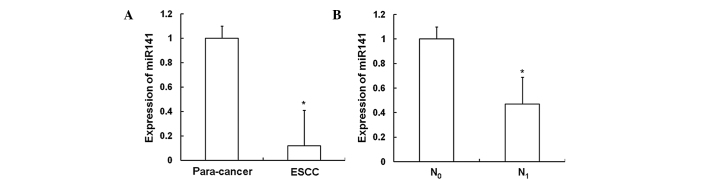
In order to investigate the expression of miR141 in ESCC tissues, RT-qPCR was performed. As shown in Fig. 1A, the expression of miR141 in ESCC tissues (0.12±0.29) was significantly decreased compared with that in the adjacent normal tissues (P<0.05). As shown in Fig. 1B, the expression level of miR141 in the tissues of patients with lymph node metastasis (N1; 0.47±0.22) was significantly decreased compared with that in the tissues of patients without such metastasis (N0; P<0.05). These results indicate that the expression of miR141 decreases as ESCC progresses.
Figure 1.
Expression of miR141 in ESCC tissues. RT-qPCR was performed to investigate the expression of miR141 in ESCC tissues. The experiments were repeated ≥3 times. Bar graphs show the expression of miR141 in (A) ESCC and adjacent normal tissues (para-cancer), as well as in (B) ESCC tissues from patients with and without lymph node metastasis. Data are presented as the mean ± standard deviation. Error bars show standard error of the mean. (A) *P<0.05 vs. adjacent normal tissue; (B) *P<0.05 vs. the lymph node metastasis-free group. ESCC, esophageal squamous cell carcinoma; miR141, microRNA-141; RT-qPCR, reverse transcription-quantitative polymerase chain reaction; N1, with lymph node metastasis; N0, without lymph node metastasis.
Expression of STAT5 is mainly localized in ESCC tissues
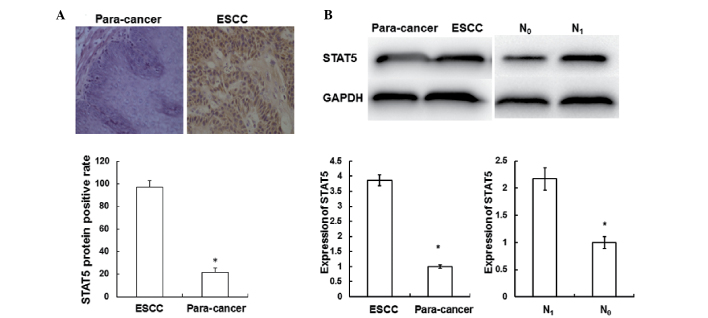
In order to investigate the distribution of STAT5 in ESCC tissues, immunohistochemical analysis was performed. As shown in Fig. 2A, STAT5 was predominantly distributed in the cytoplasm and nuclei of the ESCC tissues. The percentages of STAT5 in the ESCC tissues were significantly increased (97.2%) compared with those in the adjacent normal tissues (21.7%) (P<0.05); however, no statistically significant differences were observed in miR141 expression according to gender, age, tumor size, lesion location, differentiation and invasion (P>0.05). These results indicate that STAT5 is mainly distributed in ESCC tissues.
Figure 2.
Immunohistochemical staining and western blotting were performed to investigate the expression of STAT5 in ESCC tissues. Experiments were repeated ≥3 times. (A) Expression of STAT5 in ESCC tissues and adjacent normal tissues was detected using immunohistochemical staining. Cells with brown staining were defined as STAT5-positive (magnification, ×200). *P<0.05 vs. ESCC. (B) The expression of STAT5 in ESCC tissues, adjacent normal tissues and tissues from patients with or without lymph node metastasis was detected using western blotting. *P<0.05 for para-cancer vs. ESCC or for N0 vs. N1. GAPDH was used as a loading control. ESCC, esophageal squamous cell carcinoma; STAT5, signal transducer and activator of transcription; GAPDH, glyceraldehyde 3-phosphate dehydrogenase; N1, with lymph node metastasis; N0, without lymph node metastasis.
STAT5 protein is upregulated in ESCC tissues
In order to investigate the STAT5 protein level in ESCC tissues, western blotting was performed. As shown in Fig. 2B, when compared with the adjacent normal tissues, the protein level of STAT5 in the ESCC tissues (3.87±0.18) was significantly increased (P<0.05). The protein level of STAT5 in the tissues from patients with lymph node metastasis (2.17±0.21) was significantly increased compared with that in the tissues from patients without such metastasis (P<0.05). These results indicate that the expression of STAT5 increases as ESCC progresses.
miR141 inhibits the proliferation, migration and invasion of Eca109 cells
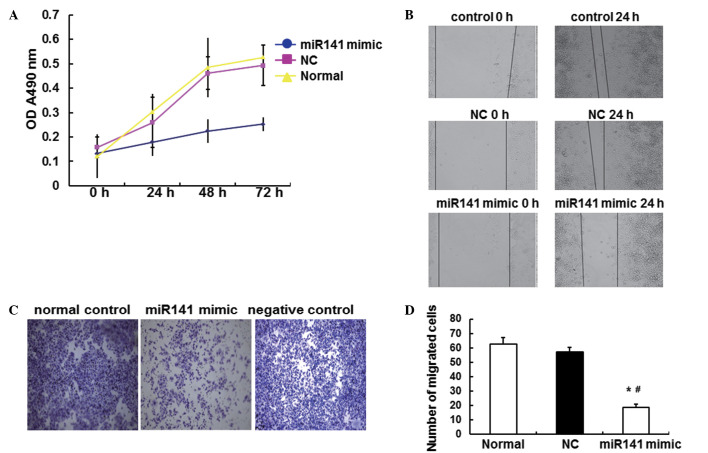
To investigate the effects of miR141 on ESCC cells, MTT, scratch and Transwell assays were performed in vitro. As shown in Fig. 3A, the proliferation of Eca109 cells in the miR141 mimic group was significantly decreased compared with that in the NC and the normal groups (P<0.05). As shown in Fig. 3B, in the scratch assay, the migration of Eca109 cells in the miR141 mimic group was significantly decreased compared with that in the NC and the normal groups (P<0.05). Furthermore, as Fig. 3C and D shows, in the Transwell assay, the invasion of Eca109 cells in the miR141 mimic group was significantly decreased compared with that in the NC and normal groups (P<0.05). These results indicate that miR141 has an important effect on Eca109 cells, and can significantly inhibit the proliferation, migration and invasion of ESCC cells.
Figure 3.
Effects of miR141 on the proliferation, migration and invasion of Eca109 cells in vitro. MTT, scratch and Transwell invasion assays were performed to investigate the effects of miR141 on Eca109 cells. The experiments were repeated ≥3 times. (A) Graph showing the proliferation of cells detected using MTT assay. Following transfection with miR141 mimic, the proliferation of Eca109 cells was detected at 0, 24, 48 and 72 h. (B) Migration of Eca109 cells was detected using a scratch assay. Following transfection with miR141 mimic, scratches were created using a sterile pipette tip and cells were incubated for 24 h. The cells in the scratch were then counted under a microscope (magnification, ×10). (C) Images showing the invasion of Eca109 cells stained with Giemsa in the Transwell assay (magnification, ×200). (D) Bar graph showing quantitative results of the Transwell assay. The number of cells that penetrated through the membrane was counted and compared among the different groups. *P<0.05 vs. the normal control group; #P<0.05 vs. the NC group. Error bars show standard error of mean. miR141, microRNA-141; OD, optical density; NC, negative control; MTT, 3-(4,5-dimethyl-2-thiazolyl)-2,5-diphenyl-2H-tetrazolium bromide.
miR141 and STAT5 are negatively associated in ESCC tissues
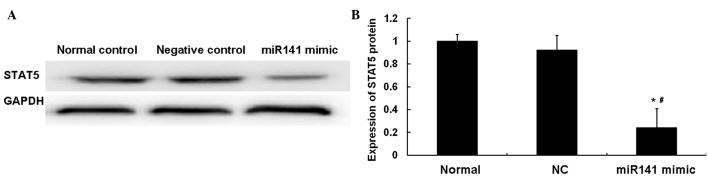
In order to investigate the association between miR141 and STAT5 in ESCC, western blotting was performed. As shown in Fig. 4, the protein level of STAT5 in the miR141 mimic group was significantly decreased compared with that in the NC group (P<0.05). No statistically significant difference was observed between the normal and NC groups (P>0.05). These results indicated that miR141 and STAT5 are negatively associated in ESCC tissues.
Figure 4.
Expression of STAT5 regulated by miR141 in Eca109 cells. Western blotting was performed to detect the expression of STAT5 in Eca109 cells transfected with miR141 mimic. Experiments were repeated ≥3 times. (A) Representative images of western blots showing STAT5 expression in Eca109 cells. (B) Bar graph showing the expression levels determined by western blotting. Expression of STAT5 in Eca109 cells was normalized to GAPDH protein expression, and compared between the groups. *P<0.05 vs. the normal control group; #P<0.05 vs. the NC group. Error bars show standard error of mean. STAT5, signal transducer and activator of transcription; miR141, microRNA-141; GAPDH, glyceraldehyde 3-phosphate dehydrogenase; NC, negative control.
Discussion
In the present study, the results indicated that miR141 functions as a tumor suppressor gene in ESCC, and that miR141 promotes the proliferation, invasion and metastasis of ESCC by decreasing the expression of the STAT5 gene.
miR141 has been reported to be abnormally expressed in various types of tumors, and miR141 has been indicated to be involved in the regulation of tumor cell proliferation, invasion and metastasis (12–14). For example, Zhou et al found that miR141 was downregulated in gastric cancer tissues and inhibited the proliferation and cell cycle of gastric cancer cells by interacting with long non-coding RNA MEG3 (12). Abedi et al found that miR141 expression was downregulated in two breast cancer cell lines, and suggested that miR141 may act through suppressing the expression and nuclear translocation of β-catenin, which is known to be important in breast cancer pathogenesis (13). In a study of endometrial carcinomas, miR141 expression was found to be upregulated, and miR141 exhibited a close association with the estrogen receptor signaling pathway (14).
In the present study, the RT-qPCR analysis showed that the expression of miR141 in the ESCC tissues was significantly decreased compared with that in the adjacent normal tissues (P<0.05). In addition, the expression of miR141 in the tissues from the patients with lymph node metastasis was significantly decreased compared with that from patients without such metastasis (P<0.05). The results of immunohistochemical analysis showed that the percentage of STAT5-positive cells in ESCC tissues was significantly increased compared with that in the adjacent normal tissues (P<0.05); however, no statistically significant differences were observed according to gender, age, differentiation, tumor size or lesion location (P>0.05). The results of western blotting showed that the expression level of STAT5 in the ESCC tissues was significantly increased compared with that in the adjacent normal tissues (P<0.05). The STAT5 expression level in the tissues from patients with lymph node metastasis was significantly increased compared with that in tissues from patients without such metastasis (P<0.05). Results of in vitro experiments showed that the proliferation, migration and invasion of Eca109 cells in the miR141 mimic group were significantly inhibited compared with those in the NC and the normal groups (P<0.05). Results of western blotting showed that the expression of STAT5 in the miR141 mimic group was significantly decreased compared with that in the NC and the normal groups (P<0.05), but there was no statistically significant difference between the NC and the normal groups (P>0.05). Furthermore, miR141 and STAT5 exhibited a negative association in the ESCC tissues.
In conclusion, the expression of miR141 was found to be significantly decreased in the ESCC tissues and to have an association with lymph node metastasis. miR141 and STAT5 were negatively associated in the ESCC tissues. The proliferation, migration and invasion of Eca109 cells were significantly inhibited by transfection with miR141 mimic in vitro, which indicates that miR141 may play an important role in the development, invasion, and metastasis of ESCC cells by regulating the expression of STAT5.
Acknowledgements
This study was supported by the Science And Technology Development Project of Linyi City (grant no. 201213022).
References
- 1.Baba Y, Watanabe M, Yoshida N, Baba H. Neoadjuvant treatment for esophageal squamous cell carcinoma. World J Gastrointest Oncol. 2014;6:121–128. doi: 10.4251/wjgo.v6.i5.121. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Roshandel G, Nourouzi A, Pourshams A, Semnani S, Merat S, Khoshnia M. Endoscopic screening for esophageal squamous cell carcinoma. Arch Iran Med. 2013;16:351–357. [PubMed] [Google Scholar]
- 3.Gu J, Wang Y, Wu X. MicroRNA in the pathogenesis and prognosis of esophageal cancer. Curr Pharm Des. 2013;19:1292–1300. doi: 10.2174/1381612811319070013. [DOI] [PubMed] [Google Scholar]
- 4.Roth C, Rack B, Müller V, Janni W, Pantel K, Schwarzenbach H. Circulating microRNAs as blood-based markers for patients with primary and metastatic breast cancer. Breast Cancer Res. 2010;12:R90. doi: 10.1186/bcr2766. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Roth C, Kasimir-Bauer S, Pantel K, Schwarzenbach H. Screening for circulating nucleic acids and caspase activity in the peripheral blood as potential diagnostic tools in lung cancer. Mol Oncol. 2011;5:281–291. doi: 10.1016/j.molonc.2011.02.002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Dorris ER, Smyth P, O'Leary JJ, Sheils O. MIR141 expression differentiates hashimoto thyroiditis from ptc and benign thyrocytes in irish archival thyroid tissues. Front Endocrinol (Lausanne) 2012;3:102. doi: 10.3389/fendo.2012.00102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Sossey-Alaoui K, Bialkowska K, Plow EF. The miR200 family of microRNAs regulates WAVE3-dependent cancer cell invasion. J Biol Chem. 2009;284:33019–33029. doi: 10.1074/jbc.M109.034553. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Severi I, Senzacqua M, Mondini E, Fazioli F, Cinti S, Giordano A. Activation of transcription factors STAT1 and STAT5 in the mouse median eminence after systemic ciliary neurotrophic factor administration. Brain Res. 2015;1622:217–229. doi: 10.1016/j.brainres.2015.06.028. [DOI] [PubMed] [Google Scholar]
- 9.Schafranek L, Nievergall E, Powell JA, Hiwase DK, Leclercq T, Hughes TP, White DL. Sustained inhibition of STAT5, but not JAK2, is essential for TKI-induced cell death in chronic myeloid leukemia. Leukemia. 2015;29:76–85. doi: 10.1038/leu.2014.156. [DOI] [PubMed] [Google Scholar]
- 10.Assefnia S, Kang K, Groeneveld S, Yamaji D, Dabydeen S, Alamri A, Liu X, Hennighausen L, Furth PA. Trp63 is regulated by STAT5 in mammary tissue and subject to differentiation in cancer. Endocr Relat Cancer. 2014;21:443–457. doi: 10.1530/ERC-14-0032. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Antoon JW, Nitzchke AM, Martin EC, Rhodes LV, Nam S, Wadsworth S, Salvo VA, Elliott S, Collins-Burow B, Nephew KP, Burow ME. Inhibition of p38 mitogen-activated protein kinase alters microRNA expression and reverses epithelial-to-mesenchymal transition. Int J Oncol. 2013;42:1139–1150. doi: 10.3892/ijo.2013.1814. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Zhou X, Ji G, Ke X, Gu H, Jin W, Zhang G. MiR-141 inhibits gastric cancer proliferation by interacting with long noncoding RNA MEG3 and down-regulating E2F3 expression. Dig Dis Sci. 2015;60:3271–3282. doi: 10.1007/s10620-015-3782-x. [DOI] [PubMed] [Google Scholar]
- 13.Abedi N, Mohammadi-Yeganeh S, Koochaki A, Karami F, Paryan M. miR-141 as potential suppressor of β-catenin in breast cancer. Tumour Biol. 2015 Jul 13; doi: 10.1007/s13277-015-3738-y. (Epub ahead of print) [DOI] [PubMed] [Google Scholar]
- 14.Dong Y, Si JW, Li WT, Liang L, Zhao J, Zhou M, Li D, Li T. miR-200a/miR-141 and miR-205 upregulation might be associated with hormone receptor status and prognosis in endometrial carcinomas. Int J Clin Exp Pathol. 2015;8:2864–2875. [PMC free article] [PubMed] [Google Scholar]