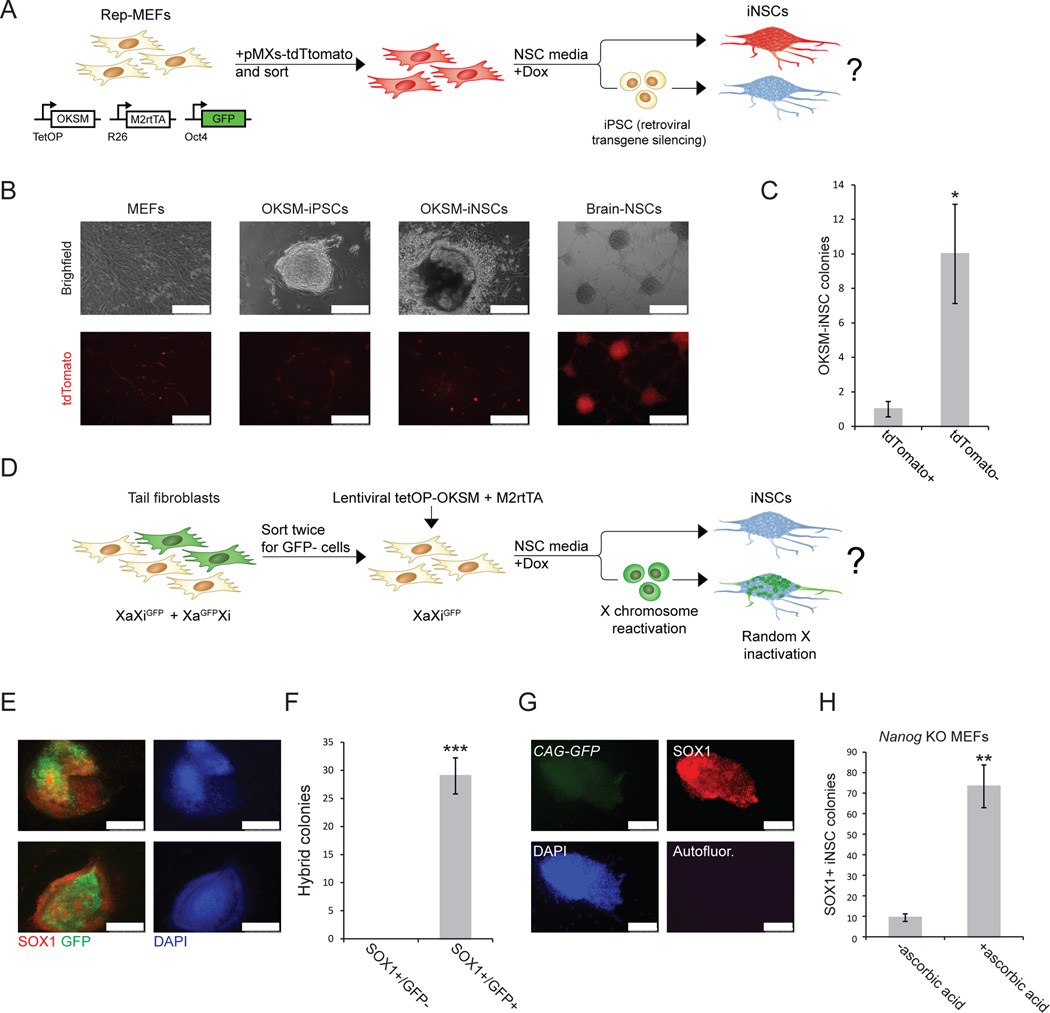
Figure 3. Silencing of retroviral transgenes and X chromosome reactivation during iNSCs induction.
(A) Experimental design to assess retroviral silencing during iNSC formation. (B) Representative images comparing tdTomato expression in MEFs, iPSCs, OKSM-iNSCs and brain-derived NSCs. Note that tdTomato+ non-reprogrammed MEFs surround the iPSC and iNSC colonies. Scale bar is 250µM. (C) Quantification of iNSC colonies expressing tdTomato+. For each replicate, 3x104 cells were used (n=5 independent replicates; error bars, s.e.m. for 5 independent experiments, *p<0.05). (D) Experimental design to follow X chromosome inactivation status during iNSC formation. Tail tip fibroblasts (TTFs) from a female mouse heterozygous for an X-linked CMV-GFP reporter (mix of XiGFPXa and XiXaGFP cells) were sorted twice for cells that carried the GFP transgene on the inactive X chromosome (XiGFPXa). These cells were reprogramed for 4–5 days in modified NSC medium, after which dox was withdrawn and GFP expression was assessed in emerging Sox1+ iNSC colonies 2–4 weeks later. (E) Representative immunofluorescence images of Sox1+ iNSC colonies that show reactivation of the silent XiGFP reporter. Scale bar is 250µM. (F) Quantification of Sox1+ iNSC colonies that reactivated the silenced X chromosome as determined by GFP reporter reactivation. All examined Sox1+ iNSC colonies showed patchy GFP expression. For each replicate, 3x104 cells were used (n=3 independent replicates; error bars, s.e.m. for 3 independent experiments, ***p<0.0005). (G) Representative immunofluorescence images showing Nanog knockout Sox1+ iNSC colony generated in the presence of ascorbate. Nanog-deficient iNSCs express the constitutive CAG-GFP marker, confirming their origin from Nanog-mutant MEFs. Scale bar is 100µM. Autofluor., autofluorescence control. (H) Quantification of Sox1+ iNSC colonies generated from Nanog-deficient MEFs with and without ascorbate. For each replicate, 3x104 cells were used (n=3 independent replicates; error bars, s.e.m. for 3 independent experiments, **p<0.005).