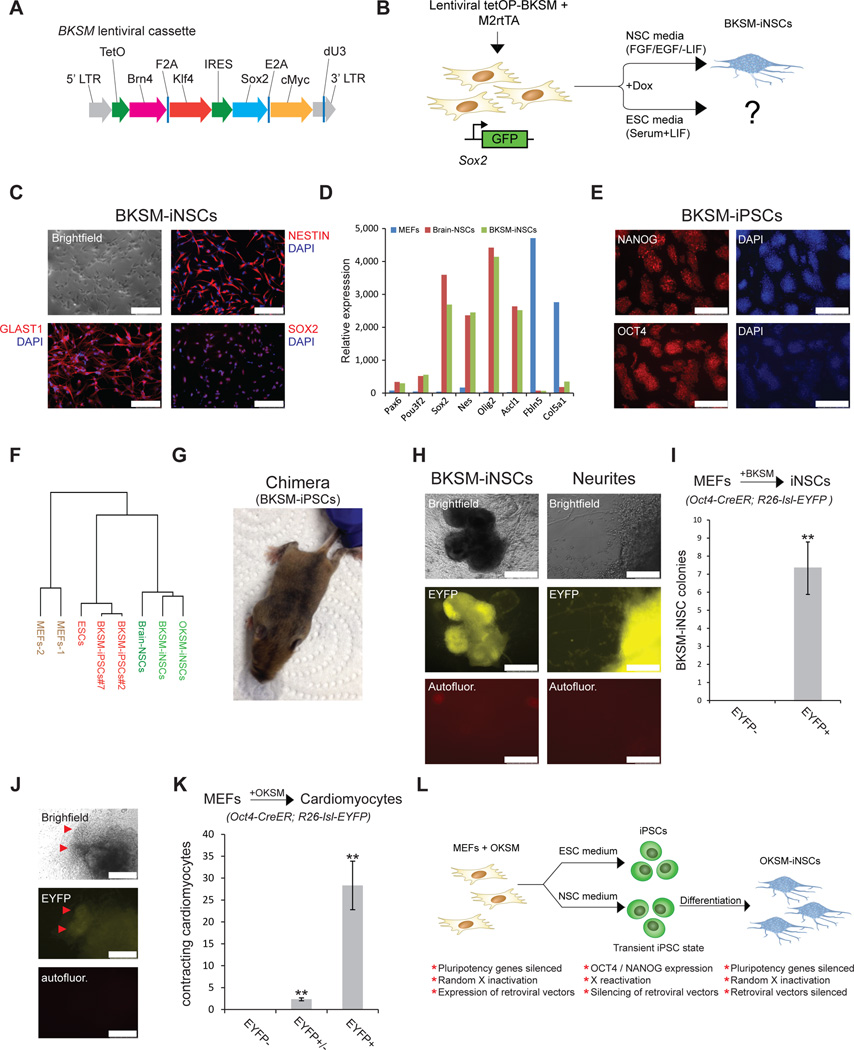
Figure 4. An alternative transdifferentiation cocktail generates iNSCs and iPSCs via an Oct4+ intermediate stage.
(A) Replacement of the Oct4 gene within the Oct4-Klf4-Sox2-cMyc (OKSM cassette) by the neuronal transcription factor Brn4 (BKSM cassette). (B) Experimental design to generate iPSCs and iNSCs using the BKSM cassette from MEFs harboring a Sox2-GFP knock-in allele in ESC or NSC culture conditions. (C) Immunofluorescence of typical NSC markers in an expanded BKSM-iNSC clone. Scale bar is 100µM. (D) Expression of selected NSC-associated genes in indicated cell lines based on microarray data. Microarray analysis was performed on expanded clonal iNSC cultures. (E) Immunofluorescence for indicated pluripotency markers in expanded BKSM-iPSC clone. Scale bar is 250µM. (F) Dendrogram based on microarray analysis of indicated samples. Note that BKSM-iNSCs and OKSM-iNSCs cluster with brain-derived NSCs while BKSM-iPSCs cluster with mouse ESCs. (G) Contribution of BKSM-iPSCs to an adult chimera as shown by agouti coat color. (H) Representative images of EYFP+ BKSM-iNSC colonies with emanating neurites generated from MEFs in conventional NSC medium. Scale bar is 500µM for iNSC colony (left) and 100µM for neurites (right). Autofluor., autofluorescence control. (I) Quantification of EYFP+ BKSM-iNSC colonies, demonstrating that all of the examined BKSM-iNSC colonies transitioned through an Oct4+ state. For each replicate, 5x105 cells were used (n=3 biological replicates; error bars, s.e.m. for 3 independent experiments, **p<0.005). (J) Representative images showing EYFP+ cells in a cluster of beating cardiomyocytes (red arrowheads). (K) Quantification of EYFP+, EYFP and EYFP+/EYFP contracting induced cardiomyocyte colonies derived from Oct4-CreER; R26-lsl-EYFP MEFs upon short OKSM expression in cardiac transdifferentiation conditions. For each replicate, 5x105 cells were used (n=3 biological replicates; error bars, s.e.m. for 3 independent experiments, **p<0.005). (L) Summary.