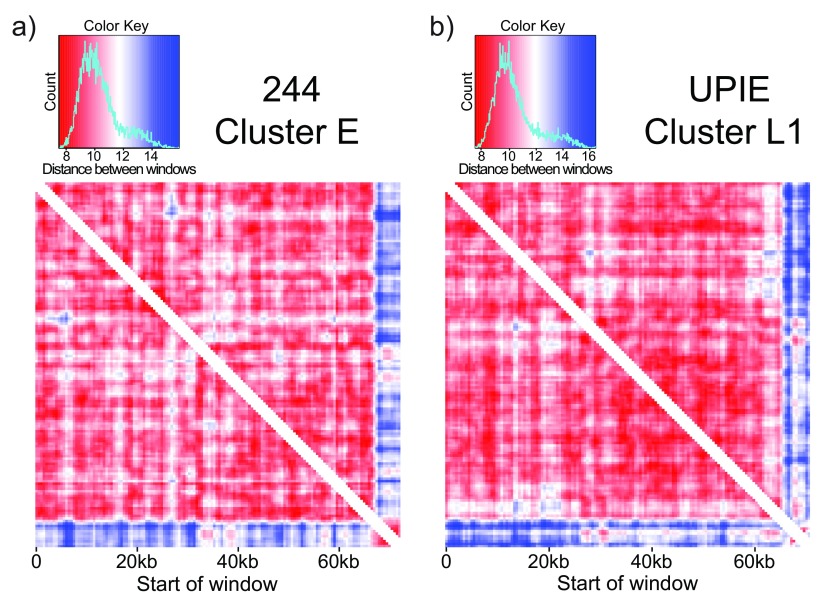
Figure 5. TUD highlights putative horizontally transferred segments.
Comparing tetranucleotide usage in a sliding window (2000bp window, 500bp step size) across phage genomes. Each entry in the heatmap is the Euclidean distance between windows. a) 244, a cluster E phage, is relatively self-similar with low distance values (red) between most windows. The last 5kb of the genome is an exception: it is self-similar but different than the rest of the genome. This signature is not driven by repetitive sequences, and represents a putative HGT event. b) UPIE, a cluster L1 phage, also has a self-similar signature at the end of the genome. However, the difference in TUD in this window is driven by two cluster of repetitive k-mers ( Figure 6).