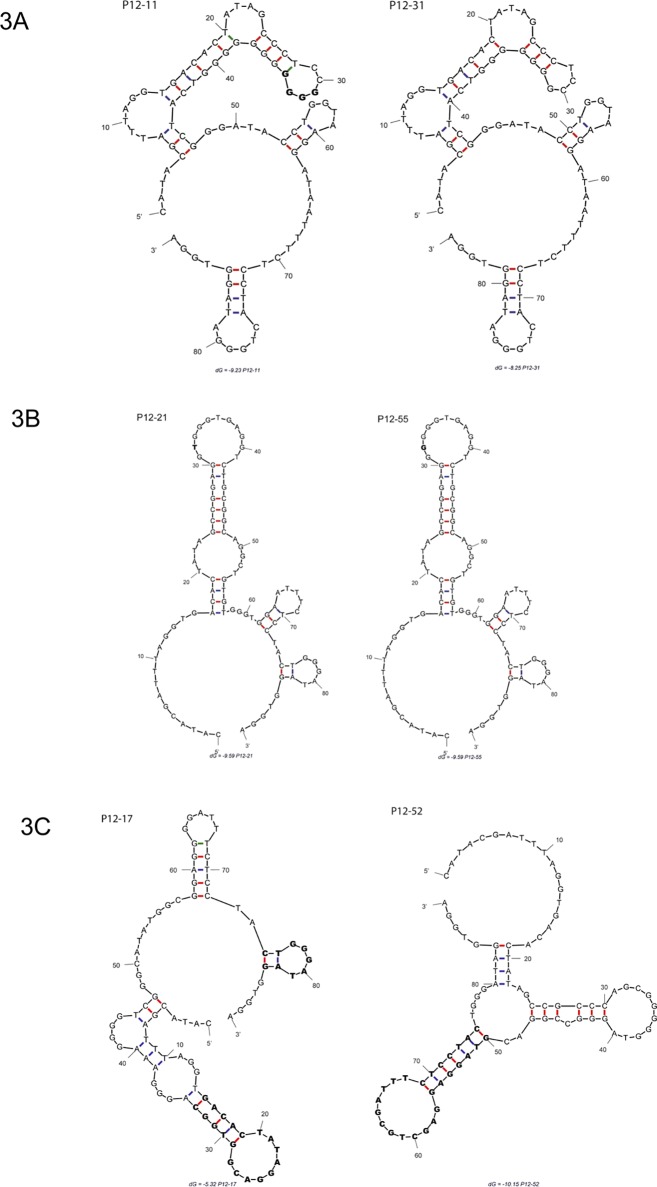
Fig 3. Prediction of pool 12 (P12) aptamer secondary structures.
The structures of aptamers with the highest E coli binding values were predicted using the mfold algorithm [63]. (A) Predicted structures of similar aptamers P12-11 and P12-31. The four G residues in P12-11 that are missing in P12-31 are depicted in bold. (B) P12-21 and P12-55 predicted structures. The different nucleotide (G or T) in the apical loop is in bold. (C) Structures of one conformer of aptamers P12-17 and P12-52, with conserved stem loops were depicted in bold.