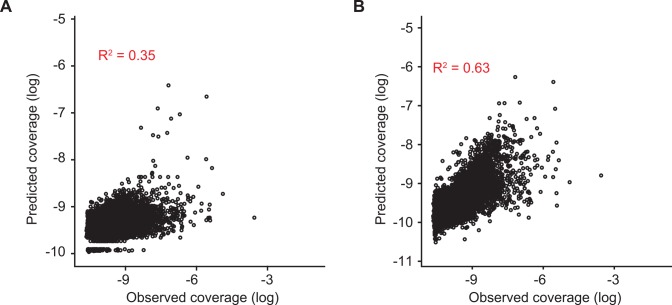
Fig 2. Genome accessibility improves prediction of ChIP-seq profiles in comparison to a model that only considers motif score.
Motif score alone explains only 35% of the observed variance (A), while the improved biophysically motivated model that incorporates genome accessibility explains 63% of the variance (B) (p<10−16, likelihood ratio test). The predicted coverage is estimated from parameters fitted for Eq 1. Coverage is represented in terms of log(pij). The panels display a subset of 10000 points that was randomly selected to reduce the density of points and improve visualization.