Figure 4.
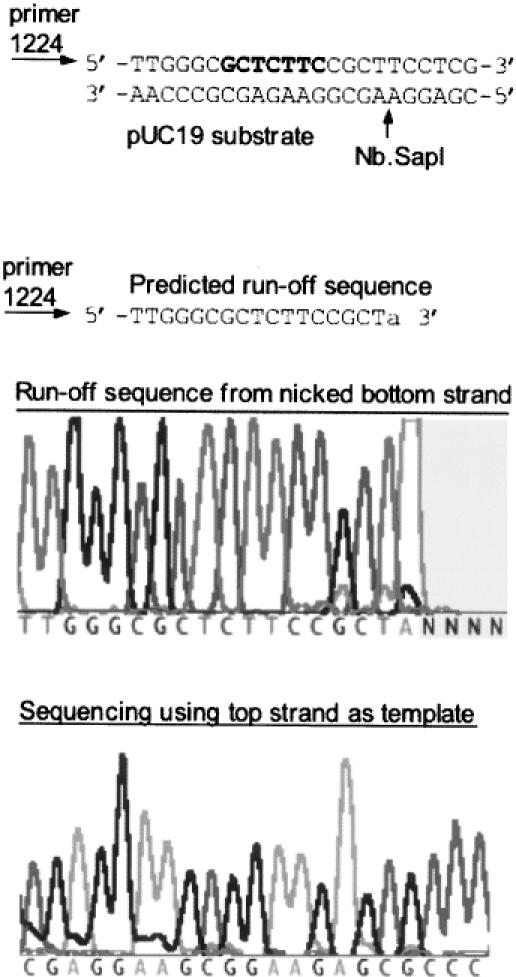
Run-off sequencing to determine the nicking site of Nb.SapI-1 (variant 33). The substrate pUC19 was incubated with purified protein for 30 min at 37°C. The nicked circular DNA product was isolated by agarose gel electrophoresis and sequenced using primers that converge on the SapI site. The sequencing reaction using primer #1224 terminated at 5′-GCTCTTCCGCTA-3′ as expected for a nicked bottom strand. AmpliTaq® DNA polymerase adds an adenine (A) at the end of the extension product (template-independent DNA transferase activity). The SapI recognition site is in bold.
